- Submissions

Full Text
Examines in Marine Biology & Oceanography
Halotolerant Bacterium used for Methane Remediation from Landfills
Jace Parkinson1, Judith L Sims2 and Ronald C Sims1*
1Biological Engineering Department, Utah State University, USA
2Utah Water Research Laboratory, Utah State University, USA
*Corresponding author: Ronald C Sims, Sustainable Waste to Bioproducts Engineering Center, Biological Engineering Department, Utah State University, Logan, Utah, USA
Submission: July 09, 2024;Published: July 29, 2024

ISSN 2578-031X Volume7 Issue2
Abstract
Methylomicrobium alcaliphilum, a halotolerant obligate methanotroph bacterium with novel biotechnology potential, was evaluated for use in biological reactors purposed for oxidizing anthropogenic methane emissions from municipal landfills. Design parameters included growth rate (μ=0.13 h-1), biomass yield on methane substrate (Yx/s=1.19gDCW/gCH4±0.02), apparent yield on substrate (Yx/s=0.49gDCW/gCH4±0.03), and specific methane degradation rate (qCH4=0.13-0.26gCH4/gDCW-h). Using these biokinetic constants for the marine bacterium and the U.S. EPA LandGEM tool to generate emission predictions for applications to the new North Valley Landfill in Utah, U.S.A., an engineered biological reactor for treatment of methane emissions was designed. A total of 250kg of M. alcaliphilum was required for treatment of 800,000 cubic meters of methane emitted from 280,000Mg of municipal landfill waste within stirred reactors at a continuous flow of 6,500L/hr sized at a total liquid phase volume of 50,000L. The applied research and testing demonstrated the potential to utilize M. alcaliphilum for mitigation of greenhouse gas emissions of methane from municipal landfills, which has been demonstrated in the literature to produce the high value bioproduct ectoine.
Keywords:Methanotrophs; Methane; Landfills; M. alcaliphilum
Introduction
Increasing atmospheric Greenhouse Gas (GHG) concentrations have necessitated development of methods for GHG treatment. CH4 is the second largest contributor to the total global atmospheric greenhouse effect. CH4 is generated as waste decomposes in landfills that released 119.8 million metric tons of carbon dioxide equivalent (MMTCO2e) of methane into the atmosphere in 2022; this represents 17 percent of the total U.S. anthropogenic methane emissions across all sectors [1]. Methanotrophs that include the halotolerant bacterium Methylomicrobium alcaliphilum are a subset of methylotrophs that assimilate methane as their carbon source. Methanotrophs produce Methane Monooxygenase (MMO), Methanol Dehydrogenase (MDH) and Formate Dehydrogenase (FDH), which allows them to utilize methane as a carbon and energy source [2]. The goal of this research was to utilize M. alcaliphilum, which has been demonstrated to produce the high value bioproduct ectoine [3], in the design of a biological reactor (fermentor) to treat methane emissions from landfills. Two specific objectives were:
i.Determine biokinetic constants for M. alcaliphilum; and
ii.Design a methane emissions treatment process for a landfill using the biokinetic constants.
Materials and Methods
M. alcaliphilum 20Z was obtained from DSMZ (Leibniz-Institut, Germany). Cryogenic stocks were stored at -72 ⁰C and prepared by adding 30% glycerol to aliquots of culture broth in a 1:1 ratio. Cultures were grown in a shaking incubator maintained at 28 ⁰C and agitated at 200rpm. A growth curve was generated by measurement of cell growth via Optical Density (OD). Analysis of Covariance (ANCOVA) was performed on the linearized data using SAS® Studio to determine the growth rate of the bacteria. Dry cell weight of the liquid cultures was determined by drying the cell pellet acquired following centrifugation of cultures.
Two treatment conditions were used to measure methane consumption rate. The first treatment used an initial headspace concentration of 80% to 20%, air to synthesized landfill gas comprised of 50% CH4 and 50% CO2. The second treatment used an initial headspace concentration of 80% to 20%, air to pure CH4. Both treatments were tested in triplicate. The gas composition of the headspace for each culture vessel was monitored by taking 2 measurements daily, one immediately before the headspace was reestablished and one immediately after. Culture absorbance measurements were acquired daily immediately after Gas Chromatography (GC) measurements of gas concentrations. Gas concentrations were determined in an Agilent 7890B GC-TCD with a GS-GasPro GC Column (60mx0.32mmx0.0μm, Agilent Part Number: 113-4362). The daily CH4 consumption was calculated by subtracting the mass of CH4 remaining from the initial mass of CH4 present. This cycle was repeated daily. The CH4 consumption was divided by time and dry weight of biomass to determine a specific CH4 consumption rate. Biological constants needed for reactor design included yield (Yx/s ), specific methane consumption rate (qCH4), and maintenance uptake rate of methane (mCH4). The relation between qCH4, μ, mCH4, and Yx/s can be explained by the Equation (1):
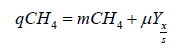
Direct calculations using experimental data were made to provide an approximation for qCH4 by dividing the mass of methane depleted in the culture vessel by the approximated average biomass in the culture vessel for the depletion period. Equation (1) takes the form of a line, with qCH4 as the dependent variable, and μ as the independent variable. By plotting qCH4 against μ, it is possible to extract mCH4 and Yx/s from the plot as the y-intercept and the inverse of the slope, respectively. A linear regression model was used to estimate the slope of this plot.
Result and Conclusion
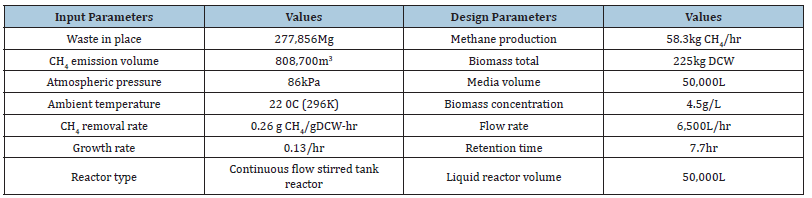
Growth rate was calculated using a semi-logarithmic transform of the plot of Optical Density (OD) with time and determining the slope of the linearized section of data. The natural logarithm transform of OD was plotted against time. Using ANCOVA with data in the time period between 2h and 17h, the growth rate was calculated as μ=0.132h-1 (standard error=0.0025, 95% confidence interval (CI) [0.127, 0.137]). Results of design parameters measured included growth rate (μ=0.13 h-1), biomass yield on substrate (Yx/s=1.19gDCW/gCH4± 0.02), apparent yield on substrate (Yx/ s=0.49gDCW/gCH4±0.03), and specific methane degradation rate (qCH4=0.13-0.26gCH4/gDCW-h). The biokinetic constants determined were applied to design a system for treating landfill emissions. A U.S. EPA Landfill Gas Emission Model (LandGEM) [4] was utilized for the new North Valley Landfill, Utah, to estimate methane emission quantity (Table 1). The reactor media volume requirement of 50,000L could be reduced through reactor designs that allow for increased biomass density and/or increased solubility of gases through sparging techniques to reduce the size of the gas bubbles, which is the target for future research.
Table 1:Input parameters and calculated values for design of the North Vallewy Landfill in Utah.
Funding
Research supported by the Utah Water Research Laboratory.
References
- EPA (2024) Inventory of U.S. greenhouse gas emissions and sinks: 1990-2022. U.S. Environmental Protection Agency, EPA 430R-24004.
- Park SY, Kim CG (2019) Application and development of methanotrophs in environmental engineering. Journal of Material Cycles and Waste Management 21(3): 415-422.
- Perez V, Molto JL, Lebrero R, Munoz R (2021) Ectoine production from biogas in waste treatment facilities. ACS Sustainable Chem Eng 9(51): 17371-17380.
- (2021) Clean Air technology center products - EPA software and manuals.
© 2024 Ronald C Sims. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






