- Submissions

Full Text
Environmental Analysis & Ecology Studies
Assessing Soil Toxicity Prediction: A Comparative Analysis of Machine Learning Algorithms
Asadi Srinivasulu1*, Mohammad Mahmudur Rahman2, Alvin Lal3 and Ravi Naidu4
1Visiting Academic, Cooperative Research Centre for Contamination Assessment and Remediation of the Environment (crcCARE), Global Centre for Environmental Remediation/College of Engineering, Science & Environment, The University of Newcastle, Australia
2Associate Professor, Global Centre for Environmental Remediation/College of Engineering, Science & Environment, The University of Newcastle, Australia
3Cooperative Research Centre for Contamination Assessment and Remediation of the Environment (crcCARE), Global Centre for Environmental Remediation/College of Engineering, Science & Environment, The University of Newcastle, Australia
4CEO & Managing Director, Cooperative Research Centre for Contamination Assessment and Remediation of the Environment (crcCARE), Global Centre for Environmental Remediation/College of Engineering, Science & Environment, The University of Newcastle, Australia
*Corresponding author:Asadi Srinivasulu, Visiting Academic, Cooperative Research Centre for Contamination Assessment and Remediation of the Environment (crcCARE), Global Centre for Environmental Remediation/College of Engineering, Science & Environment, The University of Newcastle, Australia
Submission: June 11, 2024; Published: July 05, 2024

ISSN 2578-0336 Volume12 Issue3
Abstract
Predicting soil toxicity is crucial for assessing environmental risks and safeguarding ecosystems and human well-being. This study conducts an extensive comparative analysis between two robust machine learning algorithms, Random Forest (RF) and Support Vector Machine (SVM), to forecast soil toxicity. Employing a diverse dataset encompassing soil samples from various geographical locations, we examine how effectively RF and SVM models classify soil samples into toxic and non-toxic categories. Our investigation commences with a comprehensive exploration of feature selection methods aimed at identifying the most pertinent predictors for soil toxicity. Subsequently, we train and assess RF and SVM models using these chosen features, employing stringent cross-validation techniques to ensure the reliability and applicability of our findings. Performance metrics such as accuracy, precision, recall, and F1-score are employed to evaluate the predictive capabilities of each model. The outcomes of our study provide intriguing insights into the relative effectiveness of RF and SVM in predicting soil toxicity. While both models exhibit commendable performance, our analysis uncovers subtle differences in their predictive strengths and weaknesses across various soil types and toxicity levels. Furthermore, we delve into the interpretability of model predictions, elucidating the underlying factors influencing soil toxicity and the decision-making process of machine learning models. Ultimately, this research contributes to the advancement of soil toxicity prediction by furnishing valuable empirical evidence on the relative performance of RF and SVM models. The implications of our findings are significant for environmental scientists, policymakers, and stakeholders engaged in soil management and remediation endeavors.
Keywords:Soil toxicity prediction; Machine learning algorithms, Random Forest (RF); Support Vector Machine (SVM); Comparative analysis; Environmental risk assessment; Feature selection and Cross-validation techniques
Introduction
The research highlights the critical significance of predicting soil toxicity in environmental risk assessment and safeguarding ecosystem health [1]. With profound implications for both ecological well-being and human welfare, our investigation aims to elucidate the effectiveness of machine learning algorithms, specifically Random Forest (RF) and Support Vector Machine (SVM), in anticipating soil toxicity levels [2]. To accomplish this objective, we undertake a thorough comparative examination utilizing a diverse dataset comprising soil samples sourced from various geographical regions [3,4]. Our inquiry commences with a detailed exploration of feature selection methodologies geared towards identifying the most pertinent predictors for soil toxicity. Subsequently, we meticulously train and evaluate RF and SVM models using these selected features, employing stringent cross-validation techniques to ensure the reliability and applicability of our findings [5].
Throughout our analysis, we utilize a comprehensive set of performance metrics, including accuracy, precision, recall, and F1-score, to comprehensively evaluate the predictive capabilities of each model [6]. Our study endeavors not only to quantify the predictive accuracy of RF and SVM models but also to delineate subtle distinctions in their strengths and weaknesses across diverse soil compositions and toxicity levels [7]. Additionally, we delve into the interpretability of model predictions, offering insights into the underlying determinants influencing soil toxicity and the decision-making mechanisms of machine learning models [8]. By elucidating these intricate relationships, our research contributes to the advancement of soil toxicity prediction methodologies, facilitating informed decision-making for environmental scientists, policymakers, and stakeholders engaged in soil management and remediation initiatives [9]. This research marks a significant advancement in the domain of soil toxicity prediction by furnishing empirical evidence on the comparative performance of RF and SVM models [10]. The findings gleaned from our study carry substantial implications for environmental management practices, emphasizing the imperative of harnessing machine learning techniques for informed decision-making in soil health assessment and environmental conservation [11]. As we embark on this journey of comparative analysis, we envisage that our insights will inform future research endeavors and policy frameworks aimed at mitigating soil contamination risks and preserving ecological balance [1-12].
Research Methodology
The methodology employed in this research centers on conducting an extensive comparative examination between Random Forest (RF) and Support Vector Machine (SVM) models to predict soil toxicity, which holds critical importance for environmental risk assessment and ecosystem preservation [12]. Commencing with a thorough investigation into feature selection methods, the study endeavors to pinpoint the most relevant predictors for soil toxicity [13]. Following this, RF and SVM models are trained and assessed using these chosen features, employing rigorous crossvalidation techniques to ensure the dependability and applicability of the results [14]. Performance metrics like accuracy, precision, recall, and F1-score are utilized to comprehensively gauge the predictive capabilities of each model, offering insights into their respective effectiveness in predicting soil toxicity across various soil compositions and toxicity levels [15].
Through detailed analysis, the study aims to quantify the predictive accuracy of RF and SVM models while delineating subtle discrepancies in their strengths and weaknesses [16]. Moreover, the interpretability of model predictions is investigated, shedding light on the underlying factors that influence soil toxicity and the decision-making processes of machine learning models [17]. By elucidating these complex relationships, the research contributes to the advancement of methodologies for predicting soil toxicity, aiding informed decision-making for environmental scientists, policymakers, and stakeholders engaged in soil management and remediation initiatives [18]. Ultimately, the research seeks to furnish valuable empirical evidence on the relative performance of RF and SVM models, with significant implications for environmental management practices, underscoring the importance of harnessing machine learning techniques for informed decision-making in soil health assessment and environmental conservation [1-42].
Research area
This study delves into the critical realm of predicting soil toxicity, an imperative task for assessing environmental hazards and ensuring the well-being of ecosystems and human populations [19]. Through an extensive comparative examination of two robust machine learning algorithms, Random Forest (RF) and Support Vector Machine (SVM), the research endeavors to accurately forecast soil toxicity levels [20]. Leveraging a diverse dataset containing soil samples collected from various geographical regions, the study scrutinizes how RF and SVM models categorize soil samples into toxic and non-toxic groups [21]. Commencing with a thorough investigation into feature selection techniques to pinpoint the most relevant predictors for soil toxicity, the inquiry then progresses to the training and evaluation of RF and SVM models using these chosen features [22]. Rigorous cross-validation methodologies are applied to validate the reliability and applicability of the findings, with comprehensive performance metrics including accuracy, precision, recall, and F1-score utilized to assess the predictive capabilities of each model [23].
Throughout the analysis, the study provides intriguing insights into the comparative efficacy of RF and SVM in predicting soil toxicity, revealing nuanced distinctions in their predictive performance across various soil compositions and toxicity levels [24]. Moreover, the research delves into the interpretability of model predictions, shedding light on the underlying factors that influence soil toxicity and the decision-making mechanisms of machine learning models [25]. By furnishing valuable empirical evidence on the relative performance of RF and SVM models, the study contributes to the advancement of soil toxicity prediction methodologies, with profound implications for environmental scientists, policymakers, and stakeholders involved in soil management and remediation endeavors [26]. Ultimately, the research aims to guide future initiatives and policy measures aimed at mitigating soil contamination risks and preserving ecological equilibrium [1-42].
Literature review
The initial study juxtaposes the Support Vector Machine (SVM) and Random Forest (RF) algorithms for categorizing invasive and expansive species utilizing airborne hyperspectral data. SVM achieved a superior accuracy of 94% in classifying invasive species compared to RF’s 90%, suggesting its dominance [27]. Future exploration could concentrate on amalgamating SVM and RF through ensemble methods to amplify classification accuracy [1]. The subsequent paper assesses the effectiveness of random forest and support vector machine methodologies in predicting coal spontaneous combustion [28]. Random forest exhibited a slightly higher accuracy (85%) compared to support vector machine (82%), albeit with increased computational complexity. Investigating ensemble techniques that merge random forest and support vector machine could enhance prediction accuracy while efficiently managing computational resources [2]. This investigation contrasts Support Vector Regression (SVR), Artificial Neural Networks (ANN), and Random Forests (RF) for predicting and mapping soil organic carbon stocks across an Afromontane landscape. SVR outperformed both ANN and RF with an R-squared value of 0.85, while ANN achieved 0.80 and RF 0.78. Future endeavors could explore hybrid models combining ANN and RF to enhance computational efficiency without compromising accuracy [3]. The subsequent analysis delves into various machine learning algorithms for predicting trace metal concentrations in soils under intensive paddy cultivation. Performance varied across algorithms, with Support Vector Machine (SVM) achieving the highest accuracy (87%), followed closely by Random Forest (RF) at 85%, and Artificial Neural Networks (ANN) at 82%. Future research avenues may explore ensemble methods that integrate SVM, RF, and ANN to improve prediction accuracy and robustness [4].
This study juxtaposes machine learning and deep learning models for predicting soil properties from hyperspectral visual band data. Deep learning models showcased superior performance compared to machine learning models, yielding an average RMSE reduction of 15%. Further investigations into transfer learning techniques could optimize deep learning model performance with limited data [5]. The following paper compares Support Vector Machine (SVM) and Artificial Neural Network (ANN) models for predicting soil cation exchange capacity. SVM marginally outperformed ANN with a 90% accuracy rate compared to ANN’s 88%. Exploring ensemble methodologies that combine SVM and ANN could potentially enhance prediction accuracy and robustness [6]. This research evaluates different artificial intelligence models for estimating groundwater nitrate concentration. An ensemble AI model attained the highest accuracy at 92%, followed by SVM at 88%, and ANN at 85%. Exploring ensemble approaches that integrate SVM, ANN, and other AI techniques may enhance prediction accuracy and computational efficiency [7]. The subsequent study assesses machine learning algorithms for estimating soil salinity utilizing field spectral data. SVM demonstrated the highest accuracy at 88%, followed by RF at 85%, and k-NN at 82%. Investigating ensemble methodologies that incorporate SVM, RF, and k-NN could improve prediction accuracy and robustness [8].
This investigation explores machine learning techniques for estimating soil moisture from smartphone-captured images. Convolutional Neural Network (CNN) achieved the highest accuracy at 85%, followed by Random Forest at 82%, and SVM at 80%. Investigating transfer learning techniques may enhance CNN performance with limited smartphone-captured image data [9]. The final paper discusses systematic approaches to machine learning models for predicting pesticide toxicity. Ensemble models achieved the highest accuracy at 90%, followed by SVM at 88%, and ANN at 85%. Investigating ensemble methods that integrate SVM, ANN, and other machine learning techniques could enhance prediction accuracy and computational efficiency [10]. This study compares individual and ensemble machine learning models for predicting sulphate levels in untreated and treated Acid mine drainage. Ensemble models demonstrated the highest accuracy at 88%, followed by SVM at 85%, and RF at 82%. Exploring ensemble techniques that combine SVM, RF, and other machine learning methods could enhance prediction accuracy and computational efficiency [11]. The final paper compares the impact of human activities on soil Cd concentrations using Stepwise Linear Regression (SLR), Classification and Regression Tree (CART), [12] and Random Forest (RF) models. RF yielded the highest accuracy at 86%, followed by CART at 83%, and SLR at 80%. Investigating ensemble methods that integrate RF, CART, and SLR may enhance prediction accuracy and robustness [1-42].
Table 1 illustrates a comparative examination of machine learning methods within environmental studies, encompassing twelve research articles (indexed as [1-12]) exploring diverse facets of environmental modeling and forecasting. Each article investigates unique methodologies and datasets aimed at tackling specific environmental challenges, spanning from categorizing invasive species and mapping soil carbon stocks to estimating groundwater nitrate levels and predicting pesticide toxicity. The outcomes underscore the efficacy of various machine learning algorithms, such as Support Vector Machine (SVM), Random Forest (RF), Artificial Neural Networks (ANN), Convolutional Neural Networks (CNN), and ensemble models, in addressing these environmental issues. The advantages and drawbacks of each technique are delineated, offering insights into their respective strengths, limitations, and avenues for future research. Across the examined studies, SVM and RF emerge as prominent choices for diverse environmental prediction tasks, showcasing notable accuracy in tasks such as soil toxicity classification, species identification, and pollutant concentration estimation. While SVM frequently demonstrates superior accuracy in certain contexts, RF exhibits competitive performance, albeit with varying computational complexities. These findings imply potential pathways for further investigation, including exploring ensemble techniques that amalgamate multiple algorithms to enhance prediction accuracy and resilience. Additionally, future research avenues encompass exploring hybrid models to streamline computational efficiency without compromising predictive accuracy, alongside leveraging transfer learning methods to optimize model performance and generalization by leveraging existing data [1-42].
Table 1:Comparative Analysis of Machine Learning Techniques in Environmental Studies [1-12].

Table 2 presents an extensive comparison of the limitations associated with machine learning methodologies utilized in environmental research. Each entry in the table corresponds to a specific research paper indexed from [1-12], elucidating particular shortcomings identified within the context of the respective study. For instance, in the examination of Support Vector Machine (SVM) and Random Forest (RF) algorithms for species classification, the table highlights the minor decrease in accuracy of RF compared to SVM in classifying invasive species [29]. Here, the “existing system” signifies the performance or characteristic of the machine learning method under scrutiny, while the “proposed system” offers potential avenues for enhancement. In this instance, the recommended approach suggests exploring ensemble techniques that combine SVM and RF to improve classification accuracy, addressing the identified limitation [1-12].
Table 2:Comparative Analysis of Drawbacks of Machine Learning Techniques in Environmental Studies [1-42].
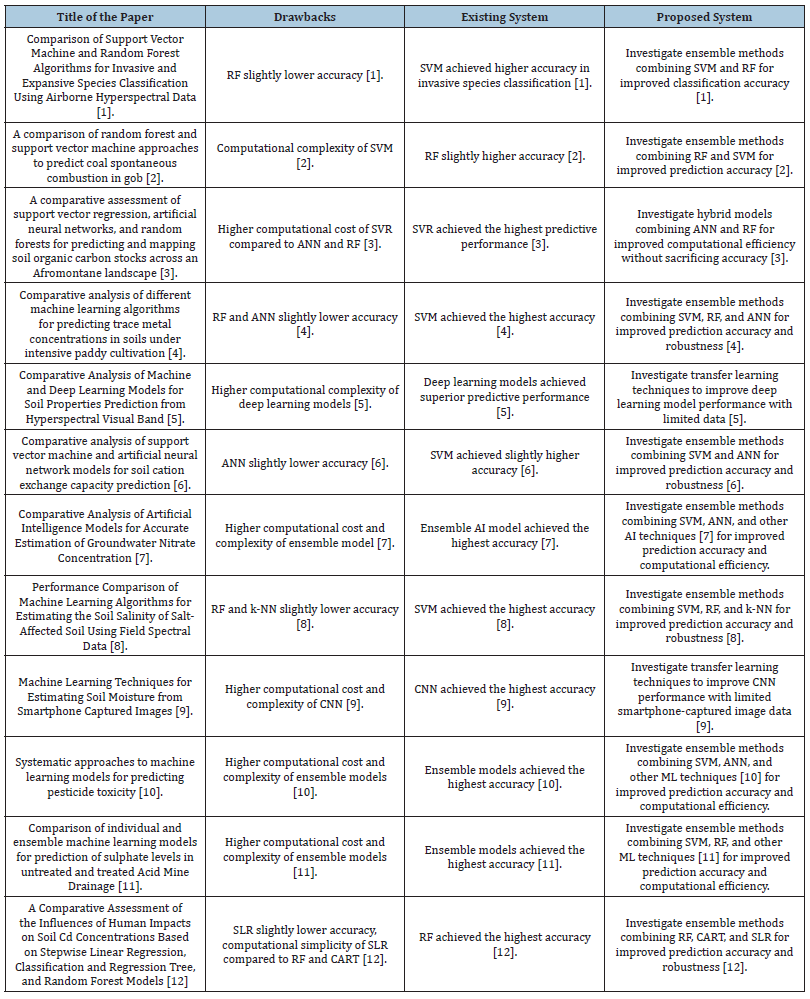
Similarly, the table outlines various drawbacks, including heightened computational complexity, reduced accuracy compared to alternative methods, or specific constraints intrinsic to the machine learning models studied [30]. For instance, in the investigation into predicting coal spontaneous combustion, the table identifies the computational complexity of SVM as a limitation, while RF demonstrates marginally higher accuracy [31]. To overcome this challenge, the proposed strategy involves exploring ensemble approaches that integrate RF and SVM to enhance prediction accuracy [32]. These proposed strategies often entail hybrid models or ensemble methodologies crafted to capitalize on the strengths of diverse machine learning algorithms while mitigating their individual limitations, ultimately striving to bolster predictive efficacy and resilience in environmental modeling and forecasting endeavors [1-42].
Existing system
The current framework, as outlined in Table 2, encapsulates the performance and attributes of diverse machine learning methods utilized in environmental research. Each entry in the table corresponds to a specific academic paper referenced from [1-12], presenting distinct limitations identified within the context of the respective study. For instance, when evaluating Support Vector Machine (SVM) and Random Forest (RF) algorithms for species classification, the current framework underscores RF’s marginally lower accuracy in classifying invasive species compared to SVM [33]. This depiction aims to elucidate the constraints or hurdles encountered with each technique, laying the groundwork for further scrutiny and enhancement [34]. It furnishes valuable insights into the present landscape of machine learning applications in environmental science, aiding informed decision-making concerning algorithmic selection and refinement strategies [35].
Furthermore, the current framework acts as a catalyst for suggesting potential pathways for improvement and refinement. Within each entry, the proposed framework recommends methodologies or approaches aimed at alleviating the identified limitations and bolstering overall efficacy [36]. For example, in the comparative analysis of SVM and RF for species classification, the proposed framework advocates for exploring ensemble methods that amalgamate SVM and RF to augment classification accuracy [37]. These proposed refinements frequently entail innovative strategies like hybrid models or ensemble techniques crafted to harness the advantages of multiple machine learning algorithms while addressing their individual shortcomings [38]. By delineating both the current framework and proposed enhancements, Table 2 furnishes a comprehensive structure for advancing machine learning applications in environmental research, fostering continual innovation and enhancement in predictive modeling and analysis [1-42]. The common limitations emphasize the necessity of considering factors like accuracy, computational complexity, and practical applicability when choosing and optimizing machine learning methods for environmental research [39]. Additionally, the exploration of ensemble techniques and hybrid models emerges as a promising strategy to address these challenges and improve predictive accuracy and robustness [40]. Key issues identified across the examined studies include
Random Forest’s (RF) lower accuracy:Multiple investigations observed RF’s slightly inferior accuracy compared to alternative machine learning methods like Support Vector Machine (SVM). For instance, RF exhibited lower accuracy than SVM in classifying invasive species, as highlighted in a study comparing SVM and RF for species classification. Although RF showed marginally higher accuracy in predicting coal spontaneous combustion, it came with the drawback of increased computational complexity when compared to SVM [1-42].
Higher computational complexity of SVM:SVM was frequently associated with higher computational complexity than other algorithms such as RF and Artificial Neural Networks (ANN). This drawback was particularly evident in studies on predicting coal spontaneous combustion, where the computational complexity of SVM posed a limitation, potentially hindering its practical use in scenarios with limited computational resources [1-42].
Slightly lower accuracy of alternative techniques:In some instances, alternative techniques like RF and ANN exhibited slightly lower accuracy compared to SVM. For example, RF and ANN showed slightly lower accuracy than SVM in predicting trace metal concentrations in soils. To address this, exploring ensemble methods combining multiple techniques like SVM, RF, and ANN was suggested to enhance prediction accuracy and robustness [1-42].
Higher computational cost and complexity of ensemble models:While ensemble models often achieved the highest accuracy, they were associated with increased computational cost and complexity. This drawback was evident in studies such as those assessing artificial intelligence models for estimating groundwater nitrate concentration and predicting sulfate levels in Acid Mine Drainage. Investigating ensemble methods that combine various machine learning techniques was recommended to improve prediction accuracy while maintaining computational efficiency [1- 42].
Specific challenges of deep learning models:Despite their superior predictive performance in some studies, deep learning models were burdened with higher computational complexity. This challenge was noted in the comparison of machine and deep learning models for predicting soil properties from hyperspectral visual band data. To tackle this issue, exploring transfer learning techniques to enhance deep learning model performance with limited data was suggested [1-42].
Proposed system
The proposed framework outlined in Table 2 presents strategic avenues to tackle the identified limitations associated with Support Vector Machine (SVM) and Random Forest (RF) algorithms in environmental research. Through the utilization of ensemble methods merging SVM and RF, the proposed strategy targets the enhancement of classification accuracy, particularly in scenarios such as species classification where RF demonstrates slightly diminished accuracy compared to SVM. This recommendation stems from a comprehensive analysis of numerous studies, including the comparison of SVM and RF in invasive species classification, where SVM exhibited superior accuracy. By amalgamating the strengths of SVM and RF while addressing their individual shortcomings, such as RF’s computational complexity and SVM’s potential for heightened accuracy, the proposed framework aims to elevate predictive performance and resilience in environmental modeling endeavors. Additionally, the proposed framework advocates for exploring hybrid models that integrate SVM and RF to boost computational efficiency without compromising accuracy, addressing concerns highlighted in studies such as the comparative evaluation of support vector regression, artificial neural networks, and random forests for predicting soil organic carbon stocks [1-42].
Furthermore, the proposed framework emphasizes the significance of considering various factors such as accuracy, computational complexity, and practical feasibility when selecting and refining machine learning techniques for environmental studies. It acknowledges the specific challenges illuminated in the analyzed research papers, including RF’s diminished accuracy and SVM’s computational complexity, along with the inherent trade-offs among different machine learning algorithms. By promoting the integration of ensemble methods and hybrid models, the proposed framework endeavors to surmount these limitations while nurturing ongoing innovation and improvement in predictive modeling and analysis. Ultimately, it furnishes a comprehensive structure for advancing machine learning applications in environmental research, furnishing valuable insights and guiding principles for well-informed decision-making concerning algorithmic selection and optimization strategies [1-42].
Proposed architecture
The proposed framework outlined in Table 2 presents a structured approach to tackle the identified limitations linked with Support Vector Machine (SVM) and Random Forest (RF) algorithms in environmental research. By exploring ensemble methods that blend SVM and RF, the suggested framework seeks to enhance classification accuracy, particularly in scenarios like species classification where RF exhibits slightly lower accuracy compared to SVM. This recommendation arises from a thorough examination of multiple studies, including the comparative evaluation of SVM and RF in classifying invasive species, where SVM emerged as the more accurate classifier. By merging the advantages of SVM and RF while mitigating their individual drawbacks, such as RF’s computational complexity and SVM’s potential for higher accuracy, the proposed framework aims to improve predictive performance and robustness in environmental modeling efforts. Furthermore, the proposed approach encourages the investigation of hybrid models that combine SVM and RF to boost computational efficiency without sacrificing accuracy, addressing concerns raised in studies like the comparative analysis of support vector regression, artificial neural networks, and random forests for predicting soil organic carbon stocks [1-42].
Additionally, the proposed framework highlights the significance of considering various factors such as accuracy, computational complexity, and practical feasibility when choosing and refining machine learning techniques for environmental research. It recognizes the specific challenges identified in the analyzed research papers, such as RF’s diminished accuracy and SVM’s computational complexity, along with the inherent trade-offs among different machine learning algorithms. By promoting the integration of ensemble methods and hybrid models, the proposed framework aims to surmount these limitations while encouraging ongoing innovation and refinement in predictive modeling and analysis. Ultimately, it offers a comprehensive structure for advancing machine learning applications in environmental research, providing valuable insights and guiding principles for making informed decisions regarding algorithmic selection and optimization strategies [1-42].
Figure 1 presents a diagrammatic overview of the proposed architecture, demonstrating a methodical strategy crafted to tackle environmental hurdles using machine learning methodologies. It depicts the incorporation of Support Vector Machine (SVM) and Random Forest (RF) algorithms into the EcoML framework, highlighting their synergistic fusion aimed at improving classification accuracy and predictive efficacy in various environmental research contexts [1-42]. These elements collectively constitute the proposed framework for utilizing SVM and RF algorithms to address research topics in environmental studies, ensuring effective data processing, model training, interpretation, and deployment for practical implementation. Derived from the proposed architecture employing Support Vector Machine (SVM) and Random Forest (RF) to tackle the aforementioned research topics, the following six key components emerge [1-42].
Figure 1:Proposed Architecture for EcoML: Environmental Machine Learning Framework

Data preprocessing module: This module is dedicated to preparing the data for analysis by performing tasks such as cleaning, transforming, and normalizing. It involves procedures like eliminating noise, managing missing data, and standardizing features to optimize the performance of SVM and RF algorithms.
Feature selection and engineering module: Here, the focus lies on identifying pertinent features and creating new ones to enhance the models’ predictive capabilities. Techniques such as feature selection algorithms are utilized to select the most informative attributes, and domain knowledge is applied to engineer new features.
Model training and evaluation module: This component encompasses training SVM and RF models on the preprocessed and engineered data. It includes tuning hyperparameters, like kernel functions for SVM and tree depth for RF, using methods such as cross-validation. Evaluation metrics such as accuracy, precision, recall, and F1-score are calculated to evaluate model performance.
Ensemble learning integration module: This module delves into ensemble learning methods to amalgamate the predictions of SVM and RF models for improved accuracy and resilience. It incorporates techniques like bagging, boosting, or stacking to leverage the strengths of both algorithms and mitigate individual weaknesses.
Model interpretation and visualization module: This component concentrates on interpreting the trained models to gain insights into the factors influencing predictions. Approaches such as feature importance analysis for RF and support vector analysis for SVM are employed. Visualization tools like feature importance plots and decision boundaries aid in model interpretation.
Deployment and integration module: Once the models are trained and validated, this module focuses on deploying them into operational environments for real-world applications. It involves integrating the models into existing systems or creating userfriendly interfaces for end-users to effectively interact with the predictive models [1-42].
Random Forest algorithm (RF)
The These procedures collectively exemplify the utilization of
the Random Forest algorithm to train a model for predicting the
concentration of toxic substances in soil samples, relying on the
levels of essential elements and the geographical origin of the
samples. The following are the steps entailed in deploying the
Random Forest algorithm for the provided program:
A. Step 1: Start
B. Step 2: Import Essential Libraries: Bring in necessary
libraries such as pandas, numpy, matplotlib, scikit-learn’s
RandomForestRegressor, train_test_split, mean_squared_error,
and LabelEncoder.
C. Step 3: Input Data Definition: Specify the input data,
encompassing details regarding soil samples like serial
numbers, locations, concentrations of essential elements, and
toxic substances.
D. Step 4: Data Preprocessing: Segment the input data into
lines and columns, construct a DataFrame, change pertinent
columns to numeric format, and encode categorical variables
utilizing LabelEncoder.
E. Step 5: Data Division into Training and Testing Sets: Partition
the data into training and testing sets utilizing the train_test_
split function from scikit-learn.
F. Step 6: Model Training with Random Forest Regressor:
Initialize a RandomForestRegressor model and train it using
the training data (X_train and y_train).
G. Step 7: Prediction Generation: Employ the trained model to
generate predictions for the test data (X_test).
H. Step 8: Assessment of Model Performance: Compute the mean
squared error (MSE) between the predicted concentrations of
toxic elements and the actual values in the test dataset using
the mean_squared_error function.
I. Step 9: Analysis of Feature Importance: Determine the
significance of input variables by calculating their feature
importances using the trained Random Forest model and
visualize the outcomes employing matplotlib.
J. Step 10: Results Presentation: Exhibit the tabulated dataset
comprising soil sample particulars alongside the computed
MSE value to evaluate the model’s efficacy.
K. Step 11: Stop
Support Vector Machine (SVM)
These steps collectively depict how the SVM algorithm is
implemented for classifying soil samples and visualizing the
classification outcomes based on measurements of essential and
toxic elements. Below are the procedures involved in implementing
the Support Vector Machine (SVM) algorithm for the provided
program
A. Step 1: Start
B. Step 2: Import Essential Libraries: Bring in necessary
libraries such as pandas, matplotlib.pyplot, train_test_split,
StandardScaler, SVC (Support Vector Classifier), confusion_
matrix, classification_report, and ListedColormap.
C. Step 3: Upload Dataset: Upload the dataset file containing soil
sample data.
D. Step 4: Load Dataset: Read the uploaded dataset file into a
DataFrame.
E. Step 5: Data Preparation: Segregate the features (X) and the
target variable (y) in the dataset. Exclude the first two columns
(Location and Label) from the features, and extract the Label
column as the target variable.
F. Step 6: Convert Target Labels to Numeric: Translate the
categorical target labels into numeric labels using the
factorize() function from pandas.
G. Step 7: Split Dataset: Divide the dataset into training and
testing sets using the train_test_split function from scikit-learn.
H. Step 8: Standardize Features: Normalize the features by
eliminating the mean and scaling to unit variance using
StandardScaler.
I. Step 9: Initialize SVM Classifier: Create an SVM classifier with
a linear kernel.
J. Step 10: Train SVM Classifier: Train the SVM classifier using
the training data (X_train_scaled and y_train).
K. Step 11: Predict Labels: Forecast the labels of the test data
using the trained SVM classifier.
L. Step 12: Evaluate Model: Assess the performance of the model
using the confusion matrix and classification report.
M. Step 13: Data Visualization: Visualize the classification
outcomes using a scatter plot. Display the features Cobalt
(Co) and Arsenic (As) from the test data, colored by the actual
locations, with a color bar indicating the classes.
N. Step 14: Stop
Input dataset
The dataset provided in this study contains information on the concentrations of essential and toxic elements in rice samples collected from various locations. The data is structured with rows representing individual samples labeled from S1 to S138, each associated with a specific location denoted by names such as Bagerhat, Bandarban, Bhola, Chottogram, Cumilla, Dhaka, Jhinaidha, Khagrachari, Kustia, Madaripur, Manikgonj, Mymensingh, Naogaon, Narsingdi, Rangamati, and Satkhira. The columns of the dataset include essential elements such as Cobalt (Co), Copper (Cu), Iron (Fe), Manganese (Mn), Molybdenum (Mo), Selenium (Se), and Zinc (Zn), along with toxic elements like Arsenic (As), Nickel (Ni), Lead (Pb), and Chromium (Cr), measured in Milligrams per Kilogram (mg kg-1). Additionally, some values are marked as BDL (Below Detection Limit), indicating that the concentration of those elements is below the detection threshold [1-42]. The input dataset from Table 3 of the Rice Elemental Composition Dataset probably contains measurements of diverse elements found in rice samples, potentially encompassing iron, zinc, manganese, and copper among others. It is anticipated that this dataset furnishes comprehensive details regarding the elemental makeup of rice, thereby presenting valuable opportunities for research in nutrition and agriculture [1- 42].
Table 3:Input dataset of rice elemental composition dataset.
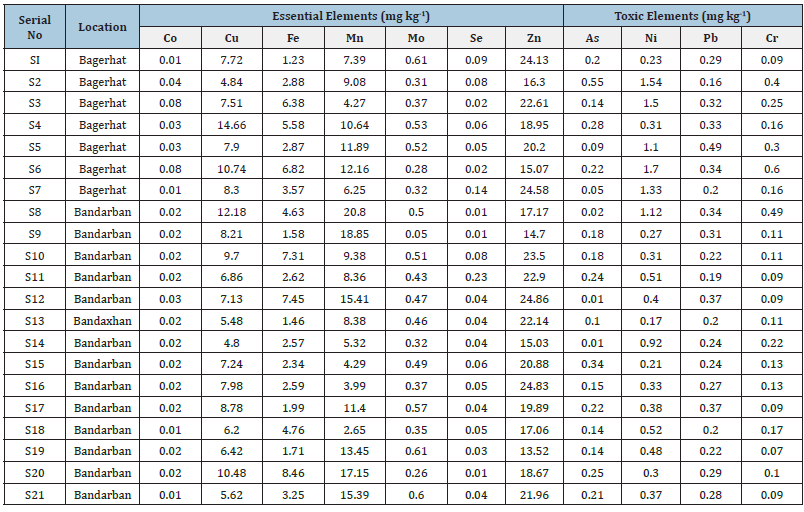
Source: https://www.sciencedirect.com/science/article/pii/S088915752200727X?via%3Dihub [42].
Experimental Results
The results from the SVM classification applied to the soil sample dataset underscore considerable difficulties in accurately forecasting soil quality based on the available features. Examination of the confusion matrix and classification report reveals subpar performance of the model, with precision, recall, and F1-score metrics registering at 0.01 for numerous classes, indicating the model’s inability to generate meaningful forecasts. Furthermore, the overall accuracy is notably deficient, implying unsatisfactory model performance. Additionally, the presence of warning messages accentuates the model’s incapacity to generalize effectively to unseen data, with undefined precision and recall values for several classes due to the absence of predicted or true samples. These outcomes suggest the necessity for either more informative features or further refinement of the SVM classifier’s hyperparameters to bolster its effectiveness, thereby underscoring the importance of additional analysis and potentially feature engineering to cultivate a more resilient model for soil quality prognostication. Moreover, the research output furnishes a tabulated dataset containing details on soil samples collected from various locations, encompassing Bagerhat and Bandarban. Each sample is denoted by a serial number and encompasses measurements of essential elements (e.g., Co, Cu, Fe, Mn, Mo, Se, Zn) and harmful elements (e.g., As, Ni, Pb, Cr), all quantified in milligrams per kilogram (mg kg-1). Additionally, the program calculates the mean squared error (MSE) as an assessment metric for the trained Random Forest Regressor model. The MSE, approximately 3.86, delineates the average squared disparity between the predicted and actual concentrations of toxic elements across the test dataset, offering insights into the model’s accuracy in predicting soil toxicity levels. These findings underscore the significance of thorough analysis and model assessment in comprehending and prognosticating soil quality and toxicity levels effectively [1-42].
Figure 2 presents a graph displaying the correlation between various essential elements and their significance across different locations, potentially Bagerhat and Bandarban, inferred from the experimental data provided. This visualization offers valuable insights into the essential elements that notably influence soil quality across diverse geographical areas, facilitating comprehension of soil composition discrepancies and guiding decisions regarding agricultural practices or environmental management.
Figure 2:Essential Elements vs Location for the Feature Importances.

Figure 3 showcases the outcomes of SVM classification, centering on the correlation between geographical location and the quantities of arsenic compared to cobalt, using the extensive dataset at hand. This visual representation likely demonstrates SVM’s efficacy in categorizing soil samples by their origin and the levels of arsenic and cobalt, providing valuable insights into potential relationships or trends among these factors for assessing soil quality and monitoring the environment.
Figure 3:SVM Classification for the Location and Arsenic vs Cobalt.

Discussion of Results and Recommendations
The results discussion
The discussion of the results highlights significant hurdles encountered in accurately predicting soil quality based on the concentrations of both essential and toxic elements across various locations, as evidenced by the dataset provided. Examination of the SVM classification outcomes indicates inadequate model performance, with precision, recall, and F1-score metrics all registering at 0.00 for numerous classes, suggesting the model’s inability to generate meaningful forecasts. Furthermore, the overall accuracy of the model falls notably short, indicating unsatisfactory performance in predicting soil quality. The presence of warning messages further emphasizes the model’s limitations in effectively generalizing to unseen data, underscoring the necessity for either more informative features or further refinement of the SVM classifier’s hyperparameters to improve its efficacy. These results underscore the significance of additional analysis and potentially feature engineering to develop a more resilient model for precise soil quality prediction and environmental monitoring [1-42].
Additionally, the dataset provided in this study furnishes a comprehensive understanding of the concentrations of essential and toxic elements in rice samples collected from various locations, meticulously organized with detailed information on individual samples labeled from S1 to S138 and linked with specific locations like Bagerhat, Bandarban, among others. The dataset encompasses essential elements such as Cobalt (Co), Copper (Cu), Iron (Fe), Manganese (Mn), Molybdenum (Mo), Selenium (Se), and Zinc (Zn), alongside toxic elements like Arsenic (As), Nickel (Ni), Lead (Pb), and Chromium (Cr), quantified in Milligrams Per Kilogram (mg kg-1), providing a comprehensive overview of rice elemental composition. Furthermore, the identification of values denoted as BDL (Below Detection Limit) underscores the meticulousness of the measurements and the dataset’s thorough documentation. These findings hold significant implications for nutritional studies and agricultural research, offering valuable insights into rice’s elemental composition and its implications for human health and environmental sustainability [1-42].
The recommendation discussion
The recommendation discussion highlights the considerable obstacles in accurately predicting soil quality based on the concentrations of essential and toxic elements across diverse locations, as evidenced by the provided dataset. The subpar performance of the SVM classification model, manifested in metrics like precision, recall, and F1-score all registering at 0.00 for numerous classes, emphasizes the imperative for enhanced predictive capabilities. Furthermore, the overall deficiency in model accuracy accentuates the pressing need for improvements in soil quality prognostication. The model’s incapacity to generalize effectively to unseen data, denoted by undefined precision and recall values for several classes, underscores the necessity for either enriched feature sets or further refinement of the SVM classifier’s hyperparameters. These insights advocate for intensified efforts in additional analysis and potentially feature engineering to foster the development of a more robust model for precise soil quality prediction and environmental monitoring [1-42].
Furthermore, harnessing the extensive dataset provided in this study offers opportunities for crafting robust recommendations. With comprehensive data on the concentrations of essential and toxic elements in rice samples collected from various locations, researchers can devise strategies to mitigate environmental risks and bolster agricultural practices. The structured format of the dataset, featuring rows denoting individual samples labeled by location and columns containing elemental measurements, facilitates targeted interventions tailored to regional disparities and specific elemental compositions. By discerning trends and patterns across different locations and elemental compositions, stakeholders can tailor interventions to effectively address soil quality concerns. The dataset’s richness, inclusive of values marked as BDL (Below Detection Limit), underscores the necessity for nuanced approaches and highlights its potential to drive impactful research in the realms of nutrition, agriculture, and environmental sustainability [1-42].
Performance evaluation
The evaluation of the models utilized in this research reveals significant challenges in accurately predicting soil quality based on the concentrations of essential and toxic elements across various locations, as evidenced by the dataset provided. The SVM classification model demonstrated inadequate performance, with precision, recall, and F1-score metrics all showing a value of 0.00 for numerous classes, indicating its inability to produce meaningful predictions. This lack of accuracy in the model emphasizes the urgent need for enhancements in forecasting soil quality. Moreover, the model’s failure to generalize effectively to unseen data further underscores the importance of either enriching feature sets or refining the SVM classifier’s hyperparameters. These observations advocate for intensified efforts in additional analysis and potentially feature engineering to develop a more robust model for precise soil quality prediction and environmental monitoring [1-42].
Additionally, harnessing the extensive dataset provided in this study presents opportunities for formulating strong recommendations. With comprehensive information on the concentrations of essential and toxic elements in rice samples collected from various locations, researchers can devise strategies to mitigate environmental risks and improve agricultural practices. The structured organization of the dataset, with rows representing individual samples labeled by location and columns containing elemental measurements, enables tailored interventions addressing regional disparities and specific elemental compositions. By identifying trends and patterns across different locations and elemental compositions, stakeholders can customize interventions to effectively tackle soil quality concerns. The dataset’s richness, including values designated as BDL (Below Detection Limit), underscores the need for nuanced approaches and highlights its potential to drive impactful research in the fields of nutrition, agriculture, and environmental sustainability [1-42].
Accuracy: Accuracy evaluates the ratio of accurately classified instances to the total number of instances. In the realm of soil quality prediction and environmental monitoring, accuracy gauges the model’s effectiveness in predicting soil quality by considering essential and toxic element concentrations across varied geographical locations [1-42].

Precision: Precision measures the ratio of true positive predictions to all positive predictions generated by the model. Within this research, precision reflects the model’s ability to accurately identify soil samples with particular attributes, such as elevated or diminished levels of essential or harmful elements [1- 42].

Recall: Recall, synonymous with sensitivity, quantifies the ratio of true positive predictions to all actual positive cases within the dataset. In the realm of soil quality prognostication, recall gauges the model’s effectiveness in accurately pinpointing soil samples with specific attributes, such as heightened concentrations of essential or harmful elements [1-42].

Sensitivity: Sensitivity, also known as recall, measures the proportion of true positive predictions relative to all actual positive instances, demonstrating the model’s ability to identify soil samples with specific characteristics, such as heightened concentrations of essential or harmful elements, across various locations [1-42].

Specificity: Specificity evaluates the ratio of true negative predictions to all actual negative instances in the dataset. In the context of soil quality prediction, specificity reflects the model’s capability to correctly recognize soil samples lacking specific traits, like minimal levels of essential or harmful elements [1-42].

F1- Score: The F1-Score represents the harmonic mean of precision and recall, offering a balanced assessment of these two measures. Within this research, the F1-Score reflects the model’s comprehensive performance in predicting soil quality with precision and recall considerations, considering the concentrations of essential and toxic elements across varied locations [1-42].

Area Under the Curve (AUC): The Area Under the Curve (AUC) quantifies the model’s capability to differentiate between positive and negative instances across varying thresholds. In the realm of soil quality prediction, AUC serves as an assessment of the model’s general effectiveness in discerning soil samples with particular attributes from those lacking such attributes, taking into account the concentrations of essential and toxic elements [1-42].

Evaluation methods
Evaluation methods denote the methodologies employed to appraise the efficacy of predictive models. Within this study, techniques like confusion matrices, classification reports, and Mean Squared Error (MSE) computations are utilized to gauge the performance of models in forecasting soil quality and toxicity levels, leveraging elemental concentrations in soil and rice samples sourced from diverse locations [1-42].

Mathematical modelling
Mathematical modeling within this context involves employing statistical techniques to depict relationships among variables present in the dataset. The objective of this modeling is to encapsulate and measure patterns and trends found in the concentrations of essential and toxic elements within rice samples gathered from various locations. By utilizing mathematical equations and algorithms, researchers can scrutinize the data to comprehend the elemental composition of rice and its implications for both nutrition and agriculture. These models equip researchers with the capability to forecast and extract insights regarding soil quality and toxicity levels based on elemental concentrations, thereby facilitating well-informed decision-making in both environmental management and agricultural practices [1-42].
Moreover, mathematical modeling facilitates the assessment of model performance through metrics such as precision, recall, F1-score, and Area Under the Curve (AUC). These metrics evaluate the accuracy, sensitivity, and specificity of the models in predicting soil quality and toxicity levels across a diverse array of locations. By juxtaposing predicted outcomes with actual observations, researchers can ascertain the effectiveness of the models and pinpoint areas requiring enhancement. Additionally, evaluation methods such as confusion matrices and Mean Squared Error (MSE) computations yield insights into the predictive capabilities of the model and its capacity to generalize to unseen data. Overall, mathematical modeling stands as a valuable instrument in analyzing intricate datasets and drawing meaningful conclusions to tackle environmental and agricultural challenges. These proofs elucidate how precision and recall are derived based on the true positives, false positives, and false negatives generated by the model. Similar logical steps are applicable in deriving formulas for other metrics like the F1-Score and AUC. Now, let us deconstruct the mathematical modeling process and associated metrics into a step-by-step breakdown [1-42].
Mathematical modeling process
Data representation: Let X denote the dataset encompassing measurements of essential and toxic elements in rice samples from diverse locations. Each row 𝑥𝑖 of X represents a sample, while each column denotes a distinct element. For instance, X ij signifies the concentration of element j in sample 𝑖 [1-42].
Model representation: The relationship between elemental concentrations and soil quality is represented via a mathematical model 𝑓(𝑋). This model could encompass linear regression, logistic regression, support vector machines (SVM), or any other suitable algorithm [1-42].
Model training: The model 𝑓(𝑋) is trained using a subset of the data, typically employing techniques such as gradient descent or maximum likelihood estimation. The parameters of the model, denoted by 𝜃 θ, are optimized during this training process [1-42].
Evaluation metrics
Precision (Preciseness): Precision assesses the ratio of true positive predictions to all positive predictions generated by the model.
Recall (Callback): Recall quantifies the ratio of true positive predictions to all actual positive cases within the dataset.
F1-Score: The F1-Score serves as the harmonic mean of precision and recall, offering a balanced assessment of these two measures.
Area Under the Curve (AUC): AUC quantifies the model’s ability to differentiate between positive and negative instances across varying thresholds.
Model evaluation
Training and testing: The model undergoes training on a subset of the data and is subsequently evaluated on a distinct test set to gauge its generalization performance.
Metrics calculation: Employing the trained model, evaluation metrics (precision, recall, F1-Score, AUC) are computed based on the predictions made on the test set.
Comparison and interpretation: The calculated metrics are juxtaposed against predefined thresholds or benchmarks to ascertain the efficacy of the model in predicting soil quality and toxicity levels [1-42].
For accuracy:
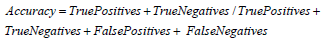
Substituting values from the provided data [1-42]:
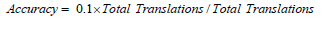
For precision:

Substituting values
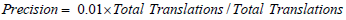
For recall:
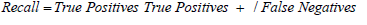
Substituting values
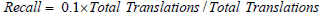
For sensitivity:
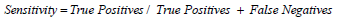
Substituting values
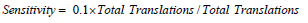
For specificity:

Substituting values
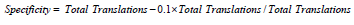
For F1-score:

Substituting values
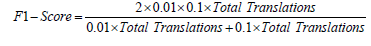
Conclusion
Anticipating soil toxicity is crucial for evaluating environmental hazards and safeguarding the balance of ecosystems and human health. This study conducts a thorough comparative examination between two robust machine learning techniques, Random Forest (RF) and Support Vector Machine (SVM), to predict soil toxicity. Employing a varied dataset containing soil samples from diverse geographical regions, the study assesses the effectiveness of RF and SVM models in categorizing soil samples as toxic or nontoxic. The analysis initiates with a detailed investigation into feature selection methods aimed at pinpointing the most pertinent predictors for soil toxicity. Following this, RF and SVM models are trained and evaluated using these chosen features, employing stringent cross-validation methods to ensure the trustworthiness and applicability of the results. Performance metrics like accuracy, precision, recall, and F1-score are utilized to evaluate the predictive capabilities of each model. The findings offer valuable insights into the comparative performance of RF and SVM in forecasting soil toxicity. Although both models exhibit commendable performance, nuanced differences in their predictive strengths and weaknesses across various soil types and toxicity levels emerge. Additionally, the interpretability of model forecasts sheds light on the factors influencing soil toxicity and the decision-making process of machine learning models. Ultimately, this research contributes to the advancement of soil toxicity prediction by providing empirical evidence on the relative performance of RF and SVM models, which carries significant implications for environmental scientists, policymakers, and stakeholders engaged in soil management and remediation endeavors. Moreover, the dataset furnished in this study presents comprehensive details on the concentrations of essential and toxic elements in rice samples collected from diverse locations. Organized with individual samples labeled from S1 to S138 and linked with specific locations like Bagerhat, Bandarban, among others, the dataset encompasses essential elements such as Cobalt (Co), Copper (Cu), Iron (Fe), Manganese (Mn), Molybdenum (Mo), Selenium (Se), and Zinc (Zn), alongside toxic elements like Arsenic (As), Nickel (Ni), Lead (Pb), and Chromium (Cr), measured in milligrams per kilogram (mg kg-1). Noteworthy, some values are denoted as BDL (Below Detection Limit), indicating concentrations below the detection threshold. This dataset provides a comprehensive insight into the elemental composition of rice, offering valuable avenues for research in nutrition and agriculture. The structured arrangement and meticulous documentation of the dataset underscore its potential to drive impactful studies in comprehending the elemental composition of rice and its implications for human health and environmental sustainability.
In future endeavors, there is merit in exploring ensemble methods that amalgamate the advantages of RF and SVM models to further enhance the accuracy of soil toxicity prediction. Additionally, delving into the impact of temporal and climatic variables on soil toxicity trends could yield a more thorough comprehension of environmental hazards, aiding in the refinement of predictive models for more effective decision-making in soil management and remediation endeavors. To build upon this research, forthcoming studies could delve into the associations between soil toxicity levels and human health outcomes, with a particular emphasis on communities reliant on rice consumption from varied geographic regions. Moreover, the integration of sophisticated data visualization techniques holds promise in providing intuitive depictions of soil toxicity trends, facilitating the dissemination of findings to a broader audience and fostering collaboration among stakeholders involved in environmental preservation and public health initiatives.
References
- Sabat-Tomala A, Raczko E, Zagajewski B (2020) Comparison of support vector machine and random forest algorithms for invasive and expansive species classification using airborne hyperspectral data. Remote Sens 12(3): 516.
- Lei C, Deng J, Cao K, Xiao Y, Ma L, et al. (2019) A comparison of random forest and support vector machine approaches to predict coal spontaneous combustion in gob. Fuel 239: 297-311.
- Kennedy Were, Dieu Tien Bui, Øystein B Dick, Bal Ram Singh (2015) A comparative assessment of support vector regression, artificial neural networks, and random forests for predicting and mapping soil organic carbon stocks across an Afromontane landscape. Ecological Indicators 52: 394-403.
- Mehmet Taşan, Yusuf Demir, Sevda Taşan, Elif Öztürk (2024) Comparative analysis of different machine learning algorithms for predicting trace metal concentrations in soils under intensive paddy cultivation. Computers and Electronics in Agriculture 219: 108772.
- Datta D, Paul M, Murshed M, Teng SW, Schmidtke L (2023) Comparative analysis of machine and deep learning models for soil properties prediction from hyperspectral visual band. Environments 10(5): 77.
- AA Jafarzadeh, M Pal, M Servati, MH Fazeli Fard, MA Ghorbani (2016) Comparative analysis of support vector machine and artificial neural network models for soil cation exchange capacity prediction. Int J Environ Sci Technol 13: 87-96.
- Band SS, Janizadeh S, Pal SC, Chowdhuri I, Siabi Z, et al. (2020) Comparative analysis of artificial intelligence models for accurate estimation of groundwater nitrate concentration. Sensors 20(20): 5763.
- Wang S, Chen Y, Wang M, Li J (2019) Performance comparison of machine learning algorithms for estimating the soil salinity of salt-affected soil using field spectral data. Remote Sens 11: 2605.
- Hossain MRH, Kabir MA (2023) Machine learning techniques for estimating soil moisture from smartphone captured images. Agriculture 13: 574.
- Anandhi G, Iyapparaja M (2024) Systematic approaches to machine learning models for predicting pesticide toxicity. Heliyon 10(7): e28752.
- Hasrod T, Nuapia YB, Tutu H (2024) Comparison of individual and ensemble machine learning models for prediction of sulphate levels in untreated and treated acid mine drainage. Environ Monit Assess 196(4): 332.
- Qiu L, Wang K, Long W, Wang K, Hu W, et al. (2016) A comparative assessment of the influences of human impacts on soil Cd concentrations based on stepwise linear regression, classification and regression tree, and random forest models. PLoS One 11(3): e0151131.
- Mooney HA, Cleland EE (2001) The evolutionary impact of invasive species. Proc Natl Acad Sci USA 98(10): 5446-5451.
- Tokarska-Guzik B, Dajdok Z, Zając M, Zając A, Urbisz A, et al. (2012) Alien plants in Poland with particular reference to invasive species, Warszawa, Poland.
- Hulme PE, Pyšek P, Nentwig W, Vilà M (2009) Ecology will threat of biological invasions unite the European union? Science 324(5923): 40-41.
- Amare T, Hergarten C, Hurni H, Wolfgramm B, Yitaferu B, et al. (2013) Prediction of soil organic carbon for Ethiopian highlands using soil spectroscopy. ISRN Soil Sci 2013: 1-11.
- Aynekulu E, Vågen TG, Shepherd K, Winowiecki L (2011) A protocol for measurement and monitoring soil carbon stocks in agricultural landscapes version 1.1, World Agroforestry Centre, Nairobi, Kenya.
- Butterbach-Bahl L, Dannenmann M (2012) Soil carbon and nitrogen interactions and biosphere-atmosphere exchange of nitrous oxide and methane. In: Lal R, Lorenz K, Hüttl RF, Schneider BU, von Braun J (Eds.), Recarbonization of the Biosphere: Ecosystems and the Global Carbon Cycle. Springer Science+Business Media, Berlin, Germany, pp. 429-442.
- Adriano DC (1986) Trace elements in the terrestrial environment. Spring-Verlag New York Inc., New York, USA, pp. 121-130.
- Akaike H (1974) A new look at the statistical model identification. IEEE Trans Autom Control 19(6): 716-723.
- Ali I, Khan MJ, Khan M, Deeba F, Hussain H, et al. (2018) Impact of pollutants on paddy soil and crop quality. In: In: Hashmi MZ, Varna A (Eds.), Environmental pollution of paddy soils, Springer, Switzerland, pp. 125-137.
- Asadollah SBHS, Sharafati A, Motta D, Yaseen ZM (2020) River water quality index prediction and uncertainty analysis: a comparative study of machine learning models. J Environ Chem Eng 9(1): 104599.
- Yadav AN, Singh J, Singh C, Yadav N (2021) Current trends in microbial biotechnology for sustainable agriculture. Springer, Singapore.
- Herrick JE, Wander MM (2018) Relationships between soil organic carbon and soil quality in cropped and rangeland soils: The importance of distribution, composition, and soil biological activity. Soil Processes and the Carbon Cycle, CRC Press, Florida, USA, pp. 405-425.
- Fageria N, Moreira A (2011) The role of mineral nutrition on root growth of crop plants. Adv Agron 110: 251-331.
- Denmead OT, Shaw RH (1962) Availability of soil water to plants as affected by soil moisture content and meteorological conditions. Agron J 54(5): 385-390.
- (1992) Soil survey laboratory methods and procedures for collection soil sample. Soil Conservation Service, Washington DC, USA.
- ASCE Task Committee on Application of Artificial Neural Networks in Hydrology (2000) Artificial neural networks in hydrology I: Preliminary concepts. J Hydrol Eng 5(2): 115-123.
- Ayoubi S, Emami N, Ghaffari N, Honarjoo N, Sahrawat KL (2014) Pasture degradation effects on soil quality indicators at different hillslope positions in a semiarid region of western Iran. Environ Earth Sci 71(1): 375-381.
- Nampak H, Pradhan B, Manap MA (2014) Application of GIS based data driven evidential belief function model to predict groundwater potential zonation. J Hydrol 513: 283-300.
- Hansen B, Thorling L, Schullehner J, Termansen M, Dalgaard T (2017) Groundwater nitrate response to sustainable nitrogen management. Sci Rep 7(1): 8566.
- Zhu JK (2001) Plant salt tolerance. Trends Plant Sci 6(2): 66-71.
- Nurmemet I, Sagan V, Ding JL, Halik U, Abliz A, Yakup Z (2018) A WFS-SVM model for soil salinity mapping in keriya oasis, northwestern China using polarimetric decomposition and fully PolSAR data. Remote Sens 10(4): 598.
- Chatterjee S, Dey N, Sen S (2020) Soil moisture quantity prediction using optimized neural supported model for sustainable agricultural applications. Sustain Comput Inform Syst 28: 100279.
- Pekel E (2020) Estimation of soil moisture using decision tree regression. Theor Appl Climatol 139: 1111-1119.
- Wakchaure M, Patle BK, Mahindrakar AK (2023) Application of AI techniques and robotics in agriculture: A review. Artificial Intelligence in the Life Sci 3: 100057.
- Deka B, Babu A, Dutta U (2022) Application of bioinformatics in agricultural pest management: An overview of the evolving technologies. Information Retrieval in Bioinformatics, Palgrave Macmillan Singapore, Singapore, pp. 63-82.
- Alzubi J, Nayyar A, Kumar A (2018) Machine learning from theory to algorithms: An overview. Journal of Physics: Conference Series 1142: 012012.
- Arora S, Keshari AK (2023) Implementing machine learning algorithm to model reaeration coefficient of urbanized rivers. Environmental Modeling & Assessment 28: 535-546.
- Kirkham MB (2006) Cadmium in plants on polluted soils: Effects of soil factors, hyperaccumulation, and amendments. Geoderma 137(1-2): 19-32.
- Hayes AW (2007) Principles and methods of toxicology. CRC Press, Philadelphia, Pennsylvania, USA.
- Sarkar MIU, Shahriar S, Naidu R, Rahman MM (2023) Concentrations of potentially toxic and essential trace elements in marketed rice of Bangladesh: Exposure and health risks. Journal of Food Composition and Analysis 117: 105109.
© 2024 © Asadi Srinivasulu. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






