- Submissions

Full Text
Significances of Bioengineering & Biosciences
Integrated RNA-Seq Analysis for Identification of Differentially Expressed Gene in Medullo-Blastoma
Sana Tabassum1, Muhammad Aizaz2*, Shahbaz Ahmad3, Mamta sharma4 and Kanika Handu5
1Department of Bioinformatics and Biotechnology, Pakistan
2Shandong Provincial Key Laboratory of Animal Resistance Biology, College of Life Sciences, Shandong Normal University, Jinan China
3Department of Microbiology, University of Swabi, Pakistan
4Project Student, India
5Intern, ESICmedical College and PGIMSR, India
*Corresponding author:Muhammad Aizaz, Shandong Provincial Key Laboratory of Animal Resistance Biology, College of Life Science, Shandong Normal University, China
Submission: April 11, 2023 Published: May 19, 2023

ISSN 2637-8078Volume6 Issue2
Abstract
Medulloblastoma is the most typical malignant brain tumor in kids. Younger children, especially babies, are at a considerably increased risk of therapeutic adverse effects. Despite significant advancement in the domain of medulloblastoma molecular biology, much remains to be accomplished in comprehending the etiology, important mechanisms causing medulloblastoma, and molecular risk stratification, as well as developing strategies of treatment with improved survival and fewer long-term squeal. So, the goal of this study is to look at and assist in the Identification and annotation of differentially expressed genes in Medulloblastoma for better treatment and long-term survival of the patients. Identification of potential therapeutic targets among the differentially expressed genes can be done by RNA-seq data analysis. This is accomplished by regulating the quality of data. We read the HTseq values with the DESeq2 tool and divided the data into normal and sick categories. This utility computes the per-exon and per-gene read counts for certain genes using RNA-seq data. We used raw numbers and a discrete distribution model to test for differential expression. Finally, annotated DESeq2 was utilized to identify differentially expressed genes. The GO enrichment and KEGG pathway in the context of CC (Cellular Component), BP (Biological Process), MF (Molecular Function), and their associated KEGG pathways indicated the functional annotation of DEGs. The analysis of the functional enrichment of DEGs revealed that the genes were engaged in oxidative Phosphorylation, Cytoplasmic Translation, and negative regulation of Ubiquitin-Protein ligase activity in terms of BP. This study will provide an overview of the current understanding of Medulloblastoma using a bioinformatics approach, as well as therapeutic prospects now being researched in surgery, radiation, and targeted medicines to maximize medulloblastoma treatment using a multidisciplinary approach.
Keywords: RNA-seq; Galaxy
Introduction
Medulloblastoma (MB) is a type of cancer that affects the Cerebellum, a part of the brain that is responsible for coordinating movement and balance. MB is a highly aggressive form of cancer that is most commonly diagnosed in babies, comprising 15-20% of all pediatric brain tumors [1,2]. It is relatively rare in adults, with most cases occurring in children between the ages of 4 and 7 [3]. Boys are more likely to develop MB than girls [4]. MB is a Heterogeneous Tumor, meaning that it has different molecular subtypes. These subtypes, which go by the names WNT, SHH, G3, and G4, have distinctive genomic landscapes and are linked to a number of risk factors [5]. Currently, the main treatment options for MB are surgery, Chemotherapy, and Radiotherapy (for children over 3 years old). Surgery involves removing as much of the tumor as possible, while Chemotherapy and Radiotherapy are used to kill cancer cells and shrink the remaining tumor. Despite these efforts, metastatic disease is still the main cause of death in MB [6], although multimodal therapy has greatly improved the prognosis for many patients. 5-year overall survival rates for MB are now more than 70%, but survivors often experience long-term neurocognitive sequelae [7]. In recent years, the public database GEO (Gene Expression Omnibus) has been widely used to analyze microarray and bioinformatics data to determine the molecular processes and underlying gene characteristics that contribute to MB and other types of cancer.
For example, the oncogene MYC is frequently altered in MB, and children with MYC-amplified MB often do not respond well to current treatment methods [8]. Bioinformatics and statistical analysis have been used to determine and predict the underlying mechanisms and pathways of MB and to develop more targeted and effective treatments for the disease [5]. These approaches involve identifying and targeting specific genes, signaling pathways, and proteins that are involved in the development and progression of MB. In current studies, we analyzed RNA-seq data to evaluate gene and transcript expression and potentially identify differentially expressed genes. The data for this analysis was obtained from the NCBI, and we used the DESeq2 program to perform differential expression analysis, normalization, and visualization. Additionally, we performed pathway analysis to identify pathways that were differentially expressed between normal and diseased samples. Our results indicated that several genes, such as those that controlled cell cycle progression and DNA damage response which is differentially expressed genes.
Materials and Methods
RNA-seq data analysis involves several steps. The first step is evaluating the quality of the readings, and if necessary, preprocessing is performed to eliminate low-quality data and artifacts. If a reference genome is available, the reads are matched to it to determine their origin. Gene and transcript expression is then measured, and novel genes and transcripts can be identified through genome-guided transcriptome assembly. Alternatively, the expression of known genes and transcripts can be measured without identifying new ones. Statistical testing may be used to examine expression differences across sample groups after abundance estimates have been derived using one of these methods.
Data retrieval
Data was downloaded from the NCBI in .gz format. The GSE189919 (from SRR17079057 to SRR17079106, there are 11 normal samples and 39 diseased samples) dataset was selected based on the experiment type (high throughput sequencing) and platform (NextSeq 550) [9].
Quality control of data
Problems with data quality can occur during the sequencing process or library preparation. These issues may include lowconfidence bases, sequence-specific bias, 5′/3′ positional bias, PCR artifacts, untrimmed adapters, and sequence contamination, which can affect mapping to reference, assembly, and expression estimation. Quality Control tools such as FastQC and PRINSEQ can be used to identify and mitigate these issues, and FastQC was used in this study to inspect the quality of the data. If low-quality bases are found at the ends of reads, they can be trimmed or a specific number of bases can be removed from either end. However, this method may also eliminate high-quality sequences. The quality of the raw files was deemed acceptable based on the FastQC report, which showed a GC percentage of 43-51% (close to the ideal range of 50-60%) and no major issues [10].
Reference alignment of reads
Alignment is the process of putting sequences together to see how and where they are connected. By matching or mapping a read to a reference genome or transcriptome, one can infer where it originated. A reference genome might be very large and contain non-unique sequences like repetitions and pseudo-genes, which reduces the ability of these regions to be mapped. Although there are millions of them and they are all short, genomes can be incredibly big. The human reference genome was aligned with query reads using bowtie aligners. The file “dna.toplevel.fa.gz,” which contains all the Chromosomes in one file, is available on the Ensemble genome browser (http://ftp.ensembl.org/pub/release-105/fasta/ homo sapiens/DNA/Homo sapiens.GRCh38.dna.toplevel.fa.gz). Then by utilizing genome FASTA files from Ensembl to assemble the HISAT2 for creating the human index, the HISAT2 program was used. For aligning the sequence with the reference index, HISAT2 is utilized. Instead of using SAM, many downstream programs now save alignments in the space-saving BAM format. The input is SAM (-S), and the output is BAM (-b). In BAM, you can read names or sort alignments by chromosomal coordinates (-n).
Reads annotation
Reads can be positioned by using genomic annotation after they have been mapped to a reference genome. Counting reads per gene, transcripts, and exon can measure gene expression and offer new choices for quality control. Although HTSeq-count is a component of Galaxy for NGS data processing. HTSeq-count stores genome annotation and aligned reads in SAM/BAM format in a GTF file. Following the exons’ gene IDs in the GTF file, Htseq-count identifies the exons which where the reads overlap and the exonlevel numbers are organized [11]. This demands that every exon of a gene have the same gene ID.
Analysis of Differential Expression (DE)
There are numerous tools for RNA-seq differential expression analysis. On data with many dimensions, differential analysis, normalization, and visualization are performed using the DESeq2 program. We used the DESeq2 tool to read the HTseq numbers and divide the data into healthy and sick groups. Using RNA-seq data, this tool determines the per-exon and per-gene read counts for certain genes. To test for differential expression, we used raw numbers and a discrete distribution model. In the end Annotate DESeq2/DEXseq was used to find the names of differentially expressed genes.
Functional enrichment analysis
The pathways and functional enrichment analysis were carried out at the functional level using DAVID (version 6.8), (a database for annotation, visualization, and integrated discovery). At three primary levels-Cellular Components (CC), Biological Processes (BP), and molecular function-the functions of anticipated DEGs were examined (MF). Later, the ggplot2 (version 3.3.0) package, an R program devoted to data visualization, was used to visualize key paths. P0.05 was regarded as statistically significant in this context.
Results
The analysis of healthy and diseased samples which were fetched from the NCBI was done thoroughly. The mega data was then processed into fastq files to make it ready as input for BAM analysis. Samples were examined for low-quality readings and adaptor contents in order to improve the quality of the analysis results. Then samples were aligned with the human reference genome.
Differentially Expressed Genes (DEGs) identification
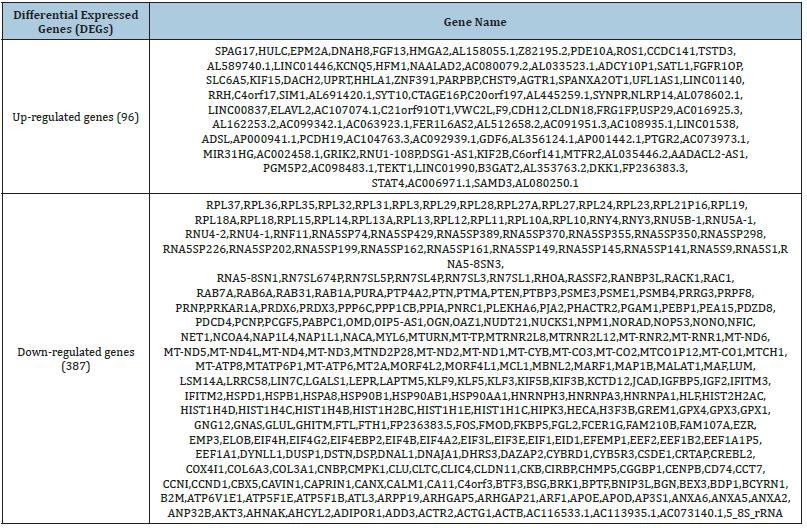
The analysis of the healthy and diseased samples which were fetched from the NCBI was done thoroughly. The mega data was then processed into fastq files to make it ready as input for SAM and BAM analysis. To map the samples with the reference human genome, high-quality reads from sample files were aligned with the reference genome. Later, the DeSeq2 tool of R was used to identify the DEGs based on log fold change (log FC), p-value, adjusted p-value, and base mean values. The term “disease” was used for diseased samples and the term “control” was used for normal samples. Differential gene expression analysis revealed a total of 387 down-regulated and 96 up-regulated genes (Table 1) while those DEGs which do not meet the criteria of logFC, and p-value were considered non-significant genes and excluded from the analysis. Moreover, Bio-conductor packages of R were used to construct a volcano plot to make a clear difference between significant and non-significant DEGs (Figure 1).
Figure 1:A volcano graphic is used to represent differentially expressed genes. Left side shows the down-regulated genes, and the right side shows the up-regulated genes, while non-significant genes were represented with central black dots.
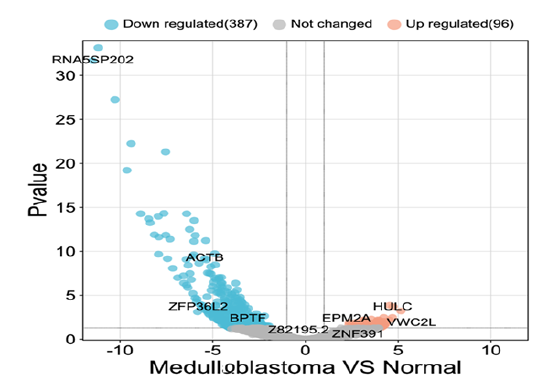
Table 1: Differential expressed genes.
In volcano plots, all the dots on the right side represented the upregulated genes, whereas the dots on the left side indicated downregulated genes. Additionally, HULC was the most significantly overexpressed up-regulated gene, and gene RNA5SP202 was the most significantly down-regulated gene with the highest expression, as demonstrated in the Volcano plot of Log2 of Fold Change and -Log10 p-values. The volcano plot also displayed the other top significant genes, including ACTB, VWC2L, and ZFP36L2.
Enrichment analysis of DEGs
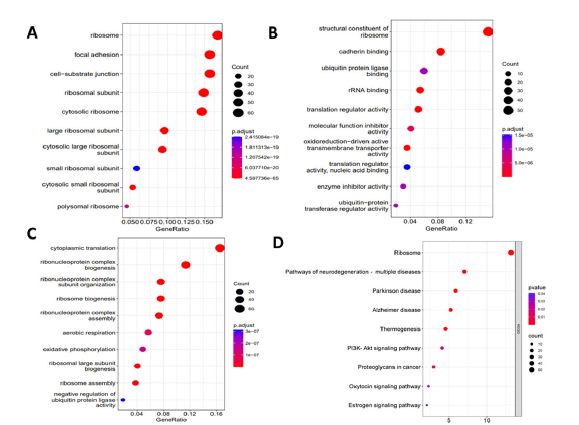
KEGG pathway and GO enrichment revealed the functional annotation of DEGs in the context of MF (Molecular Function), CC (Cellular Component), BP (Biological Process), and their associated KEGG pathways. Analysis of functional enrichment of the DEGs represented that in terms of BP, the genes were involved in oxidative phosphorylation, cytoplasmic translation, and negative regulation of ubiquitin-protein ligase activity (Figure 2). In terms of CC, genes were mainly concentrated in the ribosomal, focal adhesion, and cell-substrate adhesion (Figure 2). For the category of MF, the genes were enriched in cadherin binding, enzyme inhibitor activity, and translation regulator activity (Figure 2). KEGG pathway analysis demonstrated that genes were significantly concentrated in Parkinson’s Disease, Thermogenesis, Alzheimer’s Disease, And Proteoglycan in Cancer (Figure 2).
Figure 2:Analysis of GO enrichment. (a) Cellular Components (b) Molecular Function (c) Biological Processes (d) KEGG pathway analysis.
Discussion
While there have been significant improvements in the treatment of Medulloblastoma in recent years, including surgery, Chemotherapy, and Radiation therapy, cancer remains difficult to treat and has a relatively high mortality rate. Additionally, the treatment of Medulloblastoma can have significant side effects and long-term consequences, such as cognitive and developmental delays. As a result, there is a continued need for research and development of more effective and less toxic treatments for Medulloblastoma. In our study, we utilized RNA sequencing and bioinformatics approaches to investigate the genes, signaling pathways, and proteins involved in a disease called MB. By analyzing healthy and diseased samples, we identified several DEGs, including 387 down-regulated and 96 up-regulated genes. The most significantly up-regulated DEG was HULC, and the most significantly down-regulated DEG was gene RNA5SP202. As part of our functional enrichment analysis, we also looked at the DEGs’ functional annotation in terms of Biological Processes, Molecular Functions, Cellular Components, and KEGG pathways.
Our analysis revealed that the DEGs were involved in a variety of processes, including Oxidative Phosphorylation, Cytoplasmic Translation, Negative Regulation of Ubiquitin- Protein Ligase activity, and Ribosomal and substrate adhesion. In terms of Molecular function, the DEGs were enriched in cadherin binding, enzyme inhibitor activity, and translation regulator activity. According to KEGG pathway analysis, the DEGs were significantly enriched in pathways associated to Parkinson’s Disease, Thermogenesis, Alzheimer’s Disease, And Proteoglycan in Cancer. Previous studies have identified CDK1, WEE1, CDK2, and CCNB1 as key genes in the development of Medulloblastoma [12,13]. In medulloblastoma, VMY-1-103, a new dansylated analogue of purvalanol B, has been discovered to interfere with the advancement of Metaphase via inhibiting CDK [14]. SiRNA or the drug MK-1775 can mediate the knockdown of WEE1, a kinase involved in the G2/M cell cycle, checkpoint control, and DNA replication during the S phase, to reduce medulloblastoma cell proliferation [15]. Inhibiting WEE1 with AZD1775 has also been shown to induce apoptosis in Medulloblastoma cells [16]. WEE1 has been linked to Medulloblastoma cell Proliferation, growth, and metastasis [17]. High levels of MYC and CCNB1 expression have been identified as strong prognostic biomarkers for predicting relapse in Medulloblastoma patients [18]. CDK1 plays a significant role in neuroblastoma by partnering with CCNB1 to increase tumor cell survival [19,20]. CDK2 kinase activity has been suggested to be involved in cell proliferation, cell cycle progression, and Neuronal differentiation in medulloblastoma [21]. Targeted therapies have the potential to provide more accurate and personalized treatment for MB patients and may lead to better treatment outcomes and higher survival rates [22,23]. However, the development of targeted therapies for MB remains a challenge, as the disease is highly complex and exhibits significant Genomic Heterogeneity. This means that different molecular subtypes of MB may require different targeted therapies to be effectively treated. For example, the oncogene MYC is frequently altered in MB, and children with MYC-amplified MB often do not respond well to current treatment methods [8].
This highlights the importance of identifying targeted therapies that are specific to each molecular subtype of MB. In addition to the development of targeted therapies, research is also needed to identify new biomarkers that can be used to predict treatment response and monitor disease progression in MB patients. Identifying new biomarkers for MB could help to improve patient care by allowing doctors to monitor the effectiveness of treatment in real-time and adjust treatment plans as needed. However, the development of targeted therapies for MB remains a challenge, as the disease is highly complex and exhibits significant genomic heterogeneity. Further research is needed to fully understand the molecular mechanisms of MB and to identify the most effective targeted therapies for each molecular subtype. In addition, research is also needed to identify new biomarkers that can be used to predict treatment response and monitor disease progression in MB patients. Overall, MB is a complex and aggressive pediatric brain cancer that remains a significant challenge in the field of oncology. While progress has been made in the understanding and treatment of the disease, much more work is needed to improve patient outcomes and reduce the long-term impacts of MB.
Conclusion
The current body of research backs up a paradigm in which transcriptionally comparable tumors share clinical and molecular characteristics that will be valuable in the clinic. The discovery of molecular subgroups will almost certainly be critical in the design and execution of targeted medicines. Through this research, we identify the major genes that differ in how they are expressed in medulloblastoma. The study concluded that HULC is the highly upregulated gene, and RNA5SP202 is the highly down-regulated gene. The differentially expressed genes are functionally enriched genes.
References
- Gilbertson RJ (2004) Medulloblastoma: Signalling a change in treatment. Lancet Oncol 5(4): 209-218.
- Bartlett F, Kortmann R, Saran F (2013) Medulloblastoma. Clin Oncol (R Coll Radiol) 25(1): 36-45.
- Gerber NU (2014) Recent developments and current concepts in medulloblastoma. Cancer Treat Rev 40(3): 356-365.
- Kool M, Korshunov A, Remke M, Jones DTW, Schlanstein M, et al. (2012) Molecular subgroups of medulloblastoma: An international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol 123(4): 473-484.
- Louis DN, Perry A, Reifenberger G, Deimling AV, Figarella BD, et al. (20216) The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol 131(6): 803-820.
- Thomas A, Noël G (2019) Medulloblastoma: Optimizing care with a multidisciplinary approach. J Multidiscip Healthc 12: 335-347.
- Weishaupt H, Johansson P, Sundström A, Lubovac PZ, Olsson B, et al. (2019) Batch-normalization of cerebellar and medulloblastoma gene expression datasets utilizing empirically defined negative control genes. Bioinformatics 35(18): 3357-3364.
- Gajjar A (2020) Medulloblastoma: Improving cure rates in tandem with reduction in short-term toxicities and long-term treatment-related morbidities. Pediatr Blood Cancer 67(11): 28645.
- Lee B, Mahmud L, Pokhrel R, Murad R, Yuan M, et al. (2022) Medulloblastoma cerebrospinal fluid reveals metabolites and lipids indicative of hypoxia and cancer-specific RNAs. Acta Neuropathol Commun 10(1): 25.
- Gupta A, Gupta S, Jatawa SK, Kumar A, Suravajhala P (2019) A simplest bioinformatics pipeline for whole transcriptome sequencing: Overview of the processing and steps from raw data to downstream analysis. Bio Rxiv.
- Costa SJ, Domingues D, Lopes FM (2017) RNA-Seq differential expression analysis: An extended review and a software tool. Plos One 12(12).
- Okonechnikov K, Federico A, Schrimpf D, Sievers P, Sahm F, et al. (2023) Comparison of transcriptome profiles between medulloblastoma primary and recurrent tumors uncovers novel variance effects in relapses. Acta Neuropathologica Communications 11(1): 7.
- Northcott PA, Buchhalter I, Morrissy AS, Hovestadt V, Weischenfeldt J, et al. (2017) The whole-genome landscape of medulloblastoma subtypes. Nature 547(7663): 311-317.
- Ringer L, Sirajuddin P, Heckler M, Ghosh A, Suprynowicz F, et al. (2011) VMY-1-103 is a novel CDK inhibitor that disrupts chromosome organization and delays metaphase progression in medulloblastoma cells. Cancer Biol Ther 12(9): 818-826.
- Hirai H, Iwasawa Y, Okada M, Arai T, Nishibata T, et al. (2009) Small-molecule inhibition of wee1 kinase by MK-1775 selectively sensitizes p53-deficient tumor cells to DNA-damaging agents. Mol Cancer Ther 8(11): 2992-3000.
- Matheson CJ, Venkataraman S, Amani V, Harris PS, Backos DS, et al. (2016) A WEE1 inhibitor analog of AZD1775 maintains synergy with cisplatin and demonstrates reduced single-agent cytotoxicity in medulloblastoma cells. ACS Chem Biol 11(4): 921-930.
- Moreira DC, Venkataraman S, Subramanian A, Desisto J, Balakrishnan I, et al. (2020) Targeting MYC-driven replication stress in medulloblastoma with AZD1775 and gemcitabine. J Neurooncol 147(3): 531-545.
- Tamayo P, Cho YJ, Tsherniak A, Greulich H, Ambrogio L, et al. (2011) Predicting relapse in patients with medulloblastoma by integrating evidence from clinical and genomic features. J Clin Oncol 29(11): 1415-23.
- Islam MR, Abdulrazak LF, Alam MK, Paul BK, Ahmed K, et al. (2022) Identification of potential key genes and molecular mechanisms of medulloblastoma based on integrated bioinformatics approach. BioMed Research International.
- Rathi KS, Arif S, Koptyra M, Naqvi S, Taylor DM, et al. (2020) A transcriptome-based classifier to determine molecular subtypes in medulloblastoma. PLOS Computational Biology 16(10): 1-15.
- Guessous F, Li Y, Abounader R (2008) Signaling pathways in medulloblastoma. J Cell Physiol 217(3): 577-83.
- Shou Y, Robinson DM, Amakye DD, Rose KL, Cho YJ, et al. (2015) A five-gene hedgehog signature developed as a patient preselection tool for hedgehog inhibitor therapy in medulloblastoma. Clin Cancer Res 21(3): 585-893.
- Khatua S, Zaky W (2014) The biologic era of childhood medulloblastoma and clues to novel therapies. Future Oncol 10(4): 637-45.
© 2023 Muhammad Aizaz, This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






