- Submissions

Full Text
Research in Medical & Engineering Sciences
Genomic Analysis of BRCA2, and a Comparative Phylogenetic Analysis of BRCA2 in Humans along Different Species: A Candidate for Breast Cancer in Humans
Nimra Yaseen1, Muhammad Aetesam Nasir2, Hanzala Inam2, Izzah Mudassar2, Mamoona Arshad2, Ifrah Saroosh2, Rubace Fatima Mirza3, Syeda Zuha Naqvi4 and Muhammad Waqar Mazhar1*
1Department of Bioinformatics and Biotechnology, Government College University, Faisalabad, Pakistan
2Department of medicine and surgery, Hitec-institute of medical sciences taxila cantt, Pakistan
3Department of medicine and surgery, Wah Medical College, Wah Cantt, Pakistan
#Department of medicine and surgery, Frontiers MedicDal College, Abbottabad, Pakistan
*Corresponding author:Muhammad Waqar Mazhar, Department of Bioinformatics and Biotechnology, Government College University, Faisalabad, Pakistan
Submission: December 18, 2023;Published: January 18, 2024

ISSN: 2576-8816Volume10 Issue5
Abstract
In DNA homologous recombination repair (HRR) several proteins are encoded by BRCA1 and BRCA2. Any mutation in HRR pathway increases risk of tumor formation as a result of DNA destroying factors like salts of platinum, anthracyclines and novel agents that can damage the DNA homologous repair pathway in the form of inhibitors. Hereditary mutation in BRCA2 gene is linked with germline mutation that contribute to develop breast cancer in women. In current study, phylogenetic analysis of BRCA2 gene in 3 different species is compared to obtain similarities in that gene. Further analysis of gene structure to find exons and introns in these species is done. This proposed study supports the divergence of these 3 species from their common ancestors due to diversity in genes on chromosomes as a result of gene distribution. This current research on BRCA2 gene in humans as compared to other species help us to observe evolutionary changes in that gene through phylogenetic.
Keywords:BRCA2 gene; Germ-line mutations; Gene structures; Phylogenetic analysis
Introduction
BRCA2 gene is considered in human as a tumor suppressor gene (specifically, a responsible caretaker form of a gene), present in all humans and its protein form, is also termed as breast cancer type 2 susceptibility protein, is observed for repairing damages in DNA. BRCA2 are genes that produce such proteins to repair damaged DNA. Every organism gets two copies of these genes that are inherited from parents [1]. Sometimes BRCA2 genes are also referred as tumor suppressor genes because when they undergo mutation, they develop into cancer. When people inherit such harmful variants, they are at greater risk of several forms of cancer like ovarian and breast cancer [2].
Harmful variant can be inherited from either parent in BRCA1 and BRCA2 genes. Any mutation in any of these genes carried by child has 50% chances to develop into cancer. Such inherited mutations are termed as germline mutations that pass from parents to offspring. If a woman inherits any mutation in BRCA1 and BRCA2 genes, then the chance of ovarian and breast cancer increases in that woman’s lifetime [3].
On chromosome number 13, BRCA2 gene is present. Mutation in this gene is also passed by autosomal pattern of inheritance. As BRCA1 and BRCA2 are responsible to suppressor tumors so they have ability to control cell death and cell division [4]. Any mutation in these genes, ultimately enhance the chances of breast cancer in both men and women. Everyone inherits two copies of BRCA2 genes, one from mother and one from father. It has been observed that if a mutation occurs in one copy of gene in a person that is inherited from one parent, then still a normal copy of that gene is also present in that individual from another parent [5].
Mutation in both of these genes BRCA1 and BRCA2 causes ovarian, breast and other types of cancers. If a family undergo mutation in BRCA2 gene, then all family members show same type of mutation [6]. DNA binding part of BRCA2 gene contain 4 domains and 27 exons that consist of 82.4 kb part of genomic DNA. 11386 bp of mRNA transcript is encoded by BRCA2 gene. Site of transcription is present on 227 bp upstream region of the BRCA2 ORF on first ATG [7]. Recent research shows that only 5 to 10% cases of breast cancer develop through inherited mutation in BRCA2, while risk of breast cancer or other cancer in BRCA2 mutation depend on several other factors. Menand women carrying a mutation in BRCA2 gene are greater risk of breast, ovarian, prostate, fallopian and pancreatic cancer [8].
Material and Methods
Sequence retrieval of BRCA2 gene
Sequence of BRCA2 gene for 3 different species such as Homo sapiens, Pan troglodytes and Gorilla is retrieved from NCBI (https:// www.ncbi.nlm.nih.gov/).Protein sequence of BRCA1 was used as a query from databases. Then BLASTP was used to check the predicted protein in mammalian species including Homo sapiens, Pan troglodytes and Gorilla. Then all the conserved domains were observed for these candidate sequences by using SMART tool (http:// smart.embl-heidelberg.de/) and pfam domain (PF00170) (http://pfam.xfam.org/). Then sequences were checked to find out the BRCA2 gene in all these mammalian species. Then to predict the conserved domains of candidate gene, multiple sequence alignment was performed [9]. The full-length BRCA2 proteins from humans, chimpanzee and pan troglodytes were aligned in MEGA 7.0.21 and a phylogenetic tree was constructed that showed the sequence alignments.
Gene structure and annotations
The gene structure display server (GSDS) program (http:// gsds.cbi.pku.edu.cn/) was used to find the intron/exon regions in the gene structure of Homo sapiens, Pan troglodytes and Gorilla. Exact locations on chromosomes for the BRCA2 genes that code for BRCA2 proteins were observed from the GENE database of NCBI [10]. At the short arm telomere position to the long arm ends of telomere, the genes were organized separately on chromosome number 13 that is linked in ascending order of physical positioning. Data observed on NCBI (www.ncbi.nlm.nih.gov) was used to find all the initial GOS analysis of orthologs in three different mammalian species. The gene annotations of BRCA2 gene in humans, chimpanzee and pan troglodytes were retrieved from BLAST [11]. Each sequence of candidate specie was used as a query, to separate the BLAST searches and to form outcomes more authentic and accurate. These sequences help us to find the closer sequences and more distant sequences. After aligning all sequences, repetitive sequences were excluded from the data {Azeem, 2020 #315}.
Phylogenetic analysis of BRCA2 gene
Phylogenetic analysis of BRCA2 geneis was done to find the orthologs and homologs between human BRCA2 and two other mammalian BRCA2 genes. Using the BLAST sequence of BRCA2 genes of mammals in MEGA7, a green tree is constructed. Phylogenetic trees show evolutionary relationship between human and other mammalian species [12]. Characterization of BRCA2 gene in three different mammalian species has done by using Clustal Omega 7. By using Maximum likelihood method, we inferred the evolutionary history of BRCA2 gene in three different species. This method is based on JTT (Jones Taylor Thornton) model that is alternative model for protein alignment. The evolutionary tree is constructed according to scale with branch lengths showing evolutionary history [13].
Result
Identification of BRCA2 gene in Homo sapiens, Pan Troglodytes and Gorilla
The protein sequence of BRCA2 in candidate species was retrieved from NCBI-BLAST search. In BLAST searches, every sequence was considered as a query to obtain outcomes more precise and accurate. Through these retrieved sequences more possible matches are identified. After aligning all these sequences, repetitive sequences were removed. The signature domain of BRCA2 in these BLAST sequences was observed by using an online tool (SMART http://smart.embl-heidelberg.de) and MEME motif that showed the existence of the conserved BRCA2 domains in all candidate species (Figure 1&2).
Figure 1:Conserved BRCA2 domains a) Homo sapiens. b) Pan troglodytes. c) Gorilla gorilla.
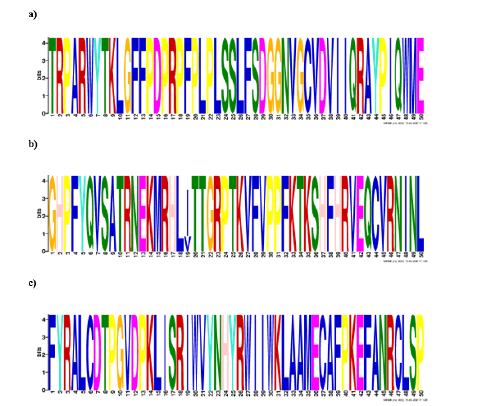
Figure 2:Motif locations of BRCA2 gene with their p-values.

Phylogenetic analysis of BRCA2 gene
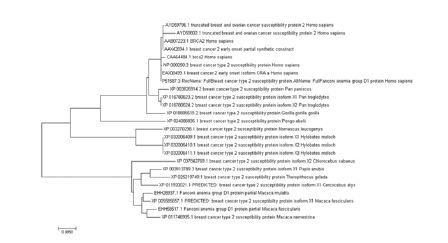
The characteristics of the evolutionary relationship between BRCA2 from Homo sapiens, Pan troglodytes and Gorilla gorilla was confirmed by the alignment of gene BRCA2 from all these species by using MEGA 7.0.21 to create rooted tree via the maximum likelihood (ML) method. Gene was found in all three candidate species. The following analyses of conserved motifs and structure of genes reinforced the characterization of BRCA2 in all species [14]. The SMART tool analysis manifested that most of the proteins of BRCA2 genes is conserved in all three candidate species while some additional domains are also present (Figure 3).
Figure 3:Phylogenetic analysis of 25 species by using MEGA-7.
Gene structure analysis of BRCA2 gene
In evolutionary analysis, the locations of introns/exons in orthologous genes are particularly well-conserved, while in paralogous genes the location of introns and exons is found approximately less conserved. In order to confirm the structural variation of BRCA2 genes, we determined the positions of introns/ exons by making comparison of CDS and genomic sequences. Although, there was substantial diversity found in the number and length of exons in all these candidate species including Homo sapiens, Pan troglodytes and Gorilla (Figures 4-6). Although, sequences retrieved at the subfamily level of these three species showed resemblance in gene structures in respect to intron number, chromosome number and exon length. Conserved motifs of BRCA2 proteins were identified and proved useful to determine the protein functions, and the additional motifs of BRCA2 gene were found functional in activating the functions of BRCA2 proteins in humans [15]. On 13 chromosome number BRCA2 gene is present in Homo sapiens. The distribution of BRCA2 gene was found dispersed on 13 chromosomes. The evolution of BRCA2 gene was further observed in all three mammalian species, while gene duplication events were examined for tandem and segmental duplications [16] (Table 1) (Figure 7).
Figure 4:Phylogenetic analysis of 25 species in circular form by using ClustalW 7.
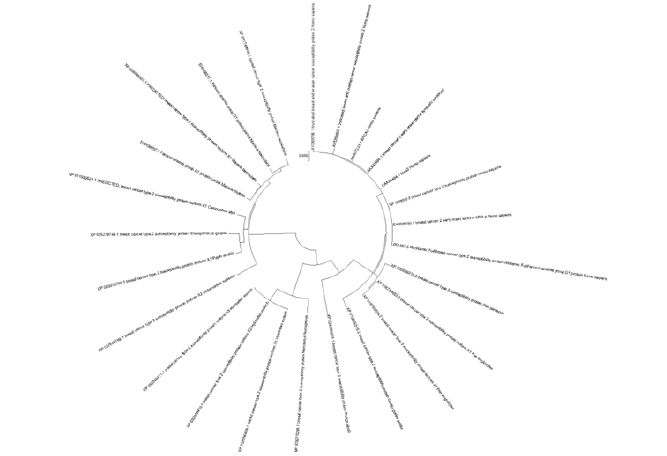
Figure 5:Phylogenetic analysis of candidate species by using MEGA-7.

Figure 6:Positions of introns and exons are determined through gene structure display server, introns are marked by black colour while exons are marked by yellow colour.
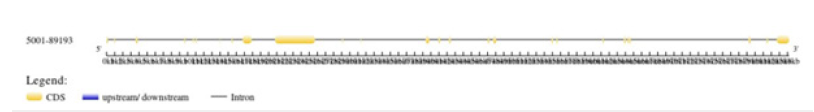
Figure 7:Graphical map of BRCA2 by using serial cloner.
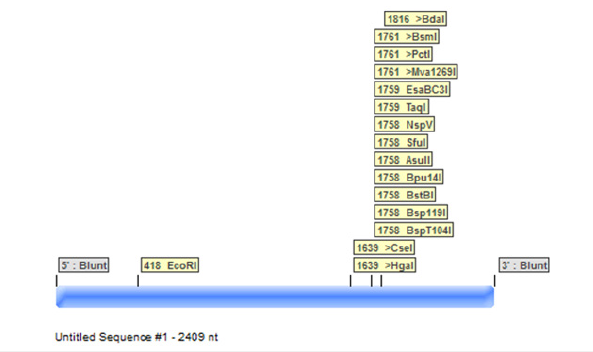
Table 1:Proposed nomenclature and important features of BRCA2gene.

Mapping of BRCA2 gene
By using serial cloner, a graphical map of BRCA2 gene is obtained. By constructing a graphical map sequence length, nucleotides and base pairs are determined. Blunt ends are generated at 5 and 3 end. In serial cloner method protein sequence of BRCA2 in FASTA format is selected and mapping of gene has done. All genes are mapped with their respective restriction sites.
Discussion
This present study shows molecular analysis of BRCA2 gene in humans and animals. It has been observed that the patients who have mutations in BRCA2 gene are at greater risk of developing cancer than the noncarriers [17]. Through current advancement in research scientists are able to edit genes by DNA repair mechanism via knock out and can directly determine the effects of mutation. Mutation carriers present in genome provide valuable information about mechanism of mutation [18]. Different characteristics of mutant genes, sequence retrieved, conserved domains, positions of introns and exons are collectively meaningful to investigate relationship of brca2 gene in mammalian species. Further, by using gene structure analysis exons count from 0 to 27 are examined precisely. In Pan troglodytes and Gorilla (western gorilla) closely related genes are observed because of their similarity in structure of introns and exons that shows their close phylogenetic relationship [19]. Results of phylogenetic tree also depict that humans, chimpanzee and western gorilla share a common ancestor. The reason behind this hypothesis is similarity in their intron exon structure. On chromosome number 13, large number of BRCA2 genes are found in all three species. That shows there is same chromosomal map for human, chimpanzee and gorilla. Subsequently, random distribution of genes on same chromosome is evidence of variation in genes. That results in an evolutionary process of three species which diverged from one another in their genome [20]. Through phylogenetic analysis, by comparing data in all three candidate species most closely related genes are observed. Hence, it is concluded that BRCA2 genes maintained their function even after divergence in humans, chimpanzee and western gorilla [21,22].
Conclusion
This research showed a high similarity of BRCA2 in structure of intron and exons present in mammalian species. Although, duplication of genes in candidate species is clearly examined. This research study depict the close evolutionary relationship between humans and chimpanzee genes as well as with western gorilla. Through this study the identified relatable genes can be used as informational source to manipulate the mutations in BRCA2 gene that are fatal.
Conflicts of Interest
The author has no conflicts of interest.
Acknowledgment
The author is thankful to GCUF, for this study.
References
- De Talhouet S, Peron J, Vuilleumier A, Friedlaender A, Viassolo V, et al. (2020) Clinical outcome of breast cancer in carriers of BRCA1 and BRCA2 mutations according to molecular subtypes. Scientific Reports 10(1): 7073.
- Peixoto A, Pinto P, Guerra J, Pinheiro M, Santos C, et al. (2020) Tumor testing for somatic and germline BRCA1/BRCA2 variants in ovarian cancer patients in the context of strong founder effects. Frontiers in Oncology 10: 1318.
- Bolton KL, Chenevix Trench G, Goh C, Sadetzki S, Ramus SJ, et al. (2012) Association between BRCA1 and BRCA2 mutations and survival in women with invasive epithelial ovarian cancer. JAMA 307(4): 382-390.
- Lou DI, McBee RM, Le UQ, Stone AC, Wilkerson GK, et al. (2014) Rapid evolution of BRCA1 and BRCA2 in humans and other primates. BMC Evolutionary Biology 14: 155.
- Kemp Z, Turnbull A, Yost S, Seal S, Mahamdallie S, et al. (2019) Evaluation of cancer-based criteria for use in mainstream BRCA1 and BRCA2 genetic testing in patients with breast cancer. JAMA Network Open 2(5): e194428.
- Hatano Y, Tamada M, Matsuo M, Hara A (2020) Molecular trajectory of BRCA1 and BRCA2 Frontiers in Oncology, 10: 361.
- Jonsson P, Bandlamudi C, Cheng ML, Srinivasan P, Chavan SS, et al. (2019) Tumour lineage shapes BRCA-mediated phenotypes. Nature 571(7766): 576-579.
- Zhao W, Wiese C, Kwon Y, Hromas R, Sung P (2019) The BRCA tumor suppressor network in chromosome damage repair by homologous recombination. Annual Review of Biochemistry 88: 221-245.
- Forbes C, Fayter D, de Kock S, Quek RG (2019) A systematic review of international guidelines and recommendations for the genetic screening, diagnosis, genetic counseling, and treatment of BRCA-mutated breast cancer. Cancer Management and Research 11: 2321-2337.
- Eccles D, Mitchell G, Monteiro A, Schmutzler R, Couch F, et al. (2015) BRCA1 and BRCA2 genetic testing-pitfalls and recommendations for managing variants of uncertain clinical significance. Annals of Oncology 26(10): 2057-2065.
- Burke LJ, Sevcik J, Gambino G, Tudini E, Mucaki EJ, et al. (2018) BRCA1 and BRCA2 5′ noncoding region variants identified in breast cancer patients alter promoter activity and protein binding. Human Mutation 39(12): 2025-2039.
- Théry JC, Krieger S, Gaildrat P, Révillion F, Buisine MP, et al. (2011) Contribution of bioinformatics predictions and functional splicing assays to the interpretation of unclassified variants of the BRCA genes. European Journal of Human Genetics 19(10): 1052- 1058.
- Walker LC, Whiley PJ, Couch FJ, Farrugia DJ, Healey S, et al. (2010) Detection of splicing aberrations caused by BRCA1 and BRCA2 sequence variants encoding missense substitutions: implications for prediction of pathogenicity. Human Mutation 31(6): E1484-E1505.
- Pfeffer CM, Ho BN, Singh AT (2017) The evolution, functions and applications of the breast cancer genes BRCA1 and BRCA2. Cancer Genomics Proteomics 14(5): 293-298.
- Lord CJ, Ashworth A (2007) RAD51, BRCA2 and DNA repair: a partial resolution. Nature Structural Molecular Biology 14(6): 461-462.
- Teulé Vega À, Lázaro García C, Blanco Guillermo I, Valle Domínguez JD, (2015) An original phylogenetic approach identified mitochondrial haplogroup T1a1 as inversely associated with breast cancer risk in BRCA2 mutation carriers. Breast Cancer Research 17(61).
- Prakash R, Zhang Y, Feng W, Jasin M (2015) Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harbor Perspectives in Biology 7(4): a016600.
- Johnston JJ, Lewis KL, Ng D, Singh LN, Wynter J, et al. (2015) Individualized iterative phenotyping for genome-wide analysis of loss-of-function mutations. The American Journal of Human Genetics 96(6): 913-925.
- Birkbak NJ, Kochupurakkal B, Izarzugaza JM, Eklund AC, Li Y, et al. (2013) Tumor mutation burden forecasts outcome in ovarian cancer with BRCA1 or BRCA2 mutations. PloS One 8(11): e80023.
- Blokzijl F, Janssen R, Van Boxtel R, Cuppen E (2018) Mutational Patterns: comprehensive genome-wide analysis of mutational processes. Genome Medicine 10(1): 33.
- Wiggins GA, Walker LC, Pearson JF (2020) Genome-wide gene expression analyses of BRCA1- and BRCA2-associated breast and ovarian tumours. Cancers 12(10): 3015.
- Acedo A, Hernández Moro C, Curiel García Á, Díez Gómez B, Velasco EA (2015) Functional classification of BRCA2 DNA variants by splicing assays in a large minigene with 9 exons. Human mutation 36(2): 210-221.
© 2023 Muhammad Waqar Mazhar. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






