- Submissions

Full Text
Research in Medical & Engineering Sciences
Escherichia coli Harboring Mcr-1 on a Novel Plasmid in a Mink in Eastern China
Xinxing Wang1, Lin Qi2* and Qiaoyun Wang2
1School of Public Health, Shandong First Medical University & Shandong Academy of Medical Sciences, China
2Therapeutic Center for Liver Disease, 960th Hospital of PLA, China
*Corresponding author: Lin Qi, Therapeutic Center for Liver Disease, 960th Hospital of PLA, China
Submission: March 22, 2022Published: March 30, 2022

ISSN: 2576-8816Volume9 Issue4
Abstract
In the current study, we described the characterization of an mcr-1-positive E. coli isolate from a dead mink in Dongping Lake, China. The molecular characterization of plasmids was also investigated. The strain was resistant to colistin, but it remained susceptible to several other agents, including amikacin, piperacillin-tazobactam, all carbapenems, and ESBLs. E. coli Mink_1 belonged to sequence type 140. Whole Genome Sequencing (WGS) showed that two plasmids, pM1mcr and pM1ctx, coexisted in the same bacterial. Blast and phylogenetic tree imply that Epidemic plasmids are vehicles for the dissemination of colistin resistance genes among the Enterobacterales. Restrictive use of antibiotics in animal husbandry should be taken to reduce the dissemination of mcr-1.
Keywords: Mcr-1; Mink; Plasmid; Escherichia coli
Introduction
The discovery of a colistin resistance gene, mcr-1, in China may herald the emergence of pandrug-resistant bacteria [1]. The gene has been found primarily in Escherichia coli (E. coli) but has also been identified in other members of the Enterobacterales, in human, animal, food, and environmental samples in over 50 countries on every continent except Antarctica [2-5]. However, the prevalence and characteristics of E.coli from mink was rarely reported. Here, we report the presence of mcr-1 in an E. coli strain cultured from a mink in Dongping Lake, China.
Methodology
E. coli Mink_1 was cultured from a dead mink which presented to a veterinary station in Dongping, China on 12th, July 2019. The strain of E. coli was isolated from liver, where bleeding spots were found. Since our research did not hurt living animals, ethical approval was not required for the study. The isolate was forwarded to SDAU for susceptibility testing. Antimicrobial Susceptibility Testing (AST) of the isolate was performed using broth dilution method. The Minimum Inhibitory Concentrations (MICs) of colistin for the E. coli isolate was also tested by the broth dilution method. The result interpretations were based on Clinical and Laboratory Standards Institute guidelines [6], except for colistin, where European Committee on Antimicrobial Susceptibility Testing guidelines breakpoints(≥2μg/mL) were used [7].
Whole Genome Sequencing (WGS) of E. coli Mink_1 was performed using a Nanopore technology and a sequence platform GridION. Complete Genome Sequencing was used to determine the location of the resistance genes. When we obtained the genome or plasmid sequences, we compared the sequences with resistance gene sequences. If the Alignment showed above 95% identity, we speculated that the resistance genes located on the responding sequence. The bioinformatics tools included Ediseq, Blastn, and Snapgene. By Blastn, we analysis the identity and coverage of our plasmid with pHNSHP45. The replicon types of plasmids were determined by PCRBased Replicon Typing (PBRT) [8-11]. The bioinformatics tools uesd include Megalign, Blastn, plasmid finder, Snapgene, etc. The sequences of mcr-1-postive plasmids was downloaded from GenBank. Then the similar sequence were cut for comparison. The construction of the phylogenetic tree was performed by the maximum likelihood method using MegAlign 8.1.4 (DNAstar Co., Ltd).
Results
Table 1Antibiotic resistance profile of E. coli Mink_1.
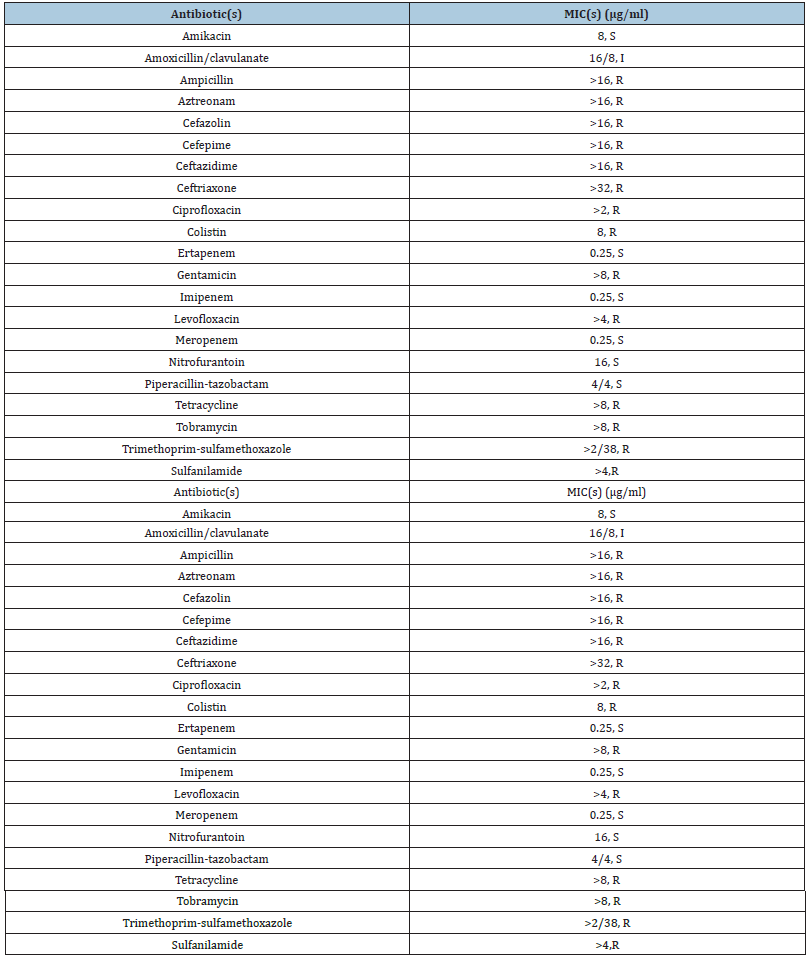
E. coli Mink_1 expressed a level of high resistance to seven different antimicrobial compounds in addition to colistin (MIC = 8mg/L) (Table 1). To note, it has a MIC of Sulfanilamide of >4μg/ mL. Susceptibility testing using double-disk test indicated an ESBL phenotype. E. coli Mink_1 belonged to sequence type 140 (ST140), a rare E. coli ST. The two plasmids, pM1mcr and pM1ctx, coexisted in the same bacterial. pM1mcr, was 238kb in size and BLAST analysis and referring to GenBank indicated that pM1mcr was identified and found to be of an IncF type that encoded a mcr-1 gene along with six additional antimicrobial resistance genes, including the ESBL gene blaCTX-M-14, and tetA, Sul2, blaTEM. The main reploins we found was FIA and FIB.
Figure 1 Phylogenetic tree of pM1mcr and similar plasmids.
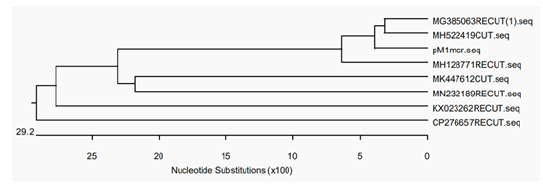
Figure 2 Structure of pM1ctx from Escherichia coli Mink 1.
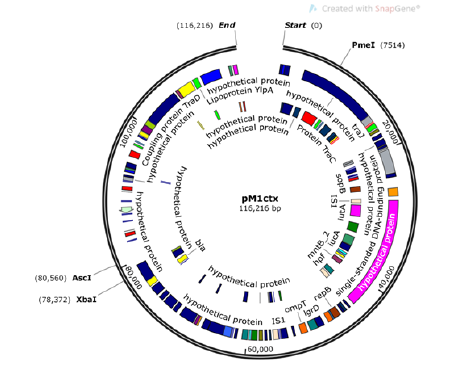
The second plasmid, pM1ctx,was 116kb in size and was assigned to IncN, which carried antibiotic resistance genes aph(3’)- Ia and blaCTX-M-1 (Figure 1). Blastn analysis showed that pM1mcr had a query coverage of 94% and maximal 99% identity to pHNSHP45, the first plasmid reported to harbor mcr-1. Results of phylogenetic tree of pM1mcr construction was shown in (Figure 2). MH522419, the most similar sequence to pM1mcr, was a strain of mcr-1- harboring Salmonella from diarrhoeal outpatients in Shanghai, China. Nucleotide sequence accession numbers. The whole genome sequence data reported in this paper have been deposited in the Genome Warehouse in National Genomics Data Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession number GWHACBG00000000 that is publicly accessible at https://bigd.big.ac.cn/gwh.
Discussion
According to the present data from GenBank, the mcr-1-positive plasmids were mainly from China, with a percentage of 52.3%, and E.coli is the primary host of mcr-1 in China. mcr-1 mainly exists in livestock breeding and animal derived food in China which varied from 1.5~30.9% in different years. He Jie detected 611 strains of Escherichia coli from 23 hospitals in 11 cities in China, and the prevalence of mcr-1 in hospital was 0.98~1.5% in hospital, which is significantly lower than that in animal husbandry, but it is worthy of people’s attention because it shows that drug-resistant genes are prevalent and spread among people. The E.coli was isolated from liver of a mink. Before the death of the mink, it showed depression and liver bleeding spots were found after autopsy. So, we speculated that the strain was pathogen, and further test should be done to determine whether the strain is pathogenic or not. The lethal pathogen may be EPEC from the environment. Meanwhile, it may also be conditional pathogen in vivo. The association between mcr- 1 and epidemic plasmids is concerning, as epidemic plasmids like IncF are vehicles for the dissemination of colisitn resistance genes among the Enterobacterales. Continued surveillance to determine the true frequency for mcr-1 is critical and measures including restrictive use of antibiotics in animal husbandry should be taken to reduce the dissemination of mcr-1.
References
- Carattoli A, Bertini A, Villa L, Falbo V, et al. (2005) Identification of plasmids by pcr-based replicon typing. Journal of Microbiological Methods 63(3): 219-228.
- Clinical and Laboratory Strandards Institute (2015) Performance standands for antimicrobial susceptibility testing; twenty-fifth information supplement M100-S25; Clinical and Laboratory Strandards Institute, Wayne, PA, USA.
- Li Zhen, Jiang W, Song C, Zhao J, Li Y, et al. (2018) Colistin resistance status of Escherichia coli and salmonella from swine and poultry and detection of mcr-1 gene. Progress in Veterinary Medicine 39(012): 20-26.
- European Committee on Antimicrobial Susceptibility Testing (EUCAST) (2016) Breakpoint tables for interpretation of MICs and zone diameters, China.
- Fernandes M, Moura Q, Sartori L, Silva K, Cunha M, et al. (2016) Silent dissemination of colistin-resistant Escherichia coli in South America could contribute to the global spread of the mcr-1 gene. Euro surveillance 21(17).
- He Jie (2018) Prevalence of colistin-resistance gene mcr-1 in Enterobacterales from blood stream infections in Zhejiang Province, China.
- Liu Y, Wang Y, Walsh R, Yi L, Zhang R, et al. (2016) Emergence of plasmid-mediated colistin resistance mechanism mcr-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infectious Disease 16(2): 161–168.
- Rapoport M, Faccone D, Pasteran F, Ceriana P, Albornoz, et al. (2016) Mcr-1-mediated colistin resistance in human infections caused by Escherichia coli: first description in Latin America. Antimicrobial Agents & Chemotherapy 60(7): 4412-4413.
- Skov R, Monnet D (2016) Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Euro Survll 21(9): 30155.
- Zeng K, Doi Y, Patil S, Huang X, Tian G (2016) Emergence of plasmid-mediated mcr-1 gene in colistin-resistant Enterobacter aerogenes and Enterobacter cloacae. Antimicrobial Agents and Chemotherapy 60(6): 3862-3863.
- Thomson K, Sanders C (1992) Detection of extended-spectrum beta-lactamases in members of the family Enterobacteriaceae: comparison of the double-disk and three-dimensional tests. Antimicrob Agents Chemother 36(9): 1877-1882.
© 2022 Lin Qi. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






