- Submissions

Full Text
Research & Development in Material Science
DNA as a Biomaterial in Diagnosis of Food Adulteration and Food Safety Assurance
Duraimurugan K1, Narendhran S2 and Manikandan M2*
1School of Community Science and Technology, Indian Institute of Engineering Science and Technology, India
2Department of Biotechnology, Sri Ramakrishna College of Arts and Science College, India
*Corresponding author: Manikandan M, Department of Biotechnology, Sri Ramakrishna College of Arts and Science College, Avinashi road, Nava India, Coimbatore- 641008, India
Submission: September 14, 2017; Published: November 30, 2017

ISSN : 2576-8840Volume2 Issue3
Abstract
Apart from general applications of DNA, it can also be used as tool to assure the food safety, which is the main aspect of this review. Increasing GMO productions also increased the concern regarding the adulteration of GMO food products in our daily life. Countries around the world, raising the regulations regarding the GMO products to the consumer's table. Subsequently, the mode of adulteration in food products with GMO products feels difficult to identify. The food products also adulterer with low cost products which can also be detected by the isolation and identification of DNA from that product. The main focus on this review is to discuss the utilization of DNA as an effective tool to detect the adulteration and GMO content in various food products like oil, honey, beverages, baked products, animal feed, fried products, adulteration of milk and milk products. In the field of food safety, microbial contamination is also a major problem. Isolation of DNA from food can also reveals the presence of microbial contamination in the earlier stage. Also, the review emphases the way of utilizing DNA in differentiating organic foods from non-organic foods.
Keywords: DNA; GMO; Adulteration; Food safety
Abbreviations: DNA: Deoxyribose Nucleic Acid; GMO: Genetically Modified Organism; GM food: Genetically Modified Food; EU: European Union; PCR: Polymerase Chain Reaction; CTAB: Cetyl Trimethyl Ammonium Bromide; AFLP: Amplified Fragment Length Polymorphism
Introduction
DNA is a genetic material of any living organism surviving on earth [1]. The pages about the nucleic acid in the history start with the Charles Darwin's first edition of "On the Origin of Species" and continued by several research people. Late to Mendel's theory on genetics, the world has turned its greatest interest towards the research about gene sciences. The period between 1950 and 2010 can be considered as golden period for the discovery and inventions in gene sciences starting from demonstration of double helix DNA topography to complete sequencing of human genome. Food and Drug Administration (FDA) has approved first genetically modified (GM) food "FLAVRSAVR Tomato" in 1994. Scientist employed the application of genetic technology in developing a hybrid variety of foods. To defeat the crisis of overpopulation rate and food demand, GMO’s acted as a solution. Belatedly, the debate on potential risk due to consumption GM food has been raised upon the people and scientists [2]. In consideration with the benefits of GM, various countries have accepted to produce GM products in their markets, whereas some of them have accepted with some restrictions [3].
The application of DNA in food is not limited only with GMO production, but also helps in various food safety measures. Since, DNA is unique for every organism in this world; it can be applied as an effective tool in the detection of food adulteration and presence of harmful microbes. As mentioned previously, some countries around the world has accepted the GMO products with various limitations like proper labeling. In order to ensure the validity of the available products in the markets, various governmental organizations conduct the screening measures at regular intervals [4-9] SYBR green-based real time PCR method was used to detect the GMO food products in markets of Kuwait shown positive results, indicating the need of stringent rules and regulations in order to protect the consumer right’s [5-7]. LightCycler-GMO screening kit based method was utilized to identify the positive GMO products sold in Saudi Arabia markets which indicates nearly 10% of them are GMO products. The major part was occupied by corn and corn based foodstuffs accounting for about 60% of total GMO products [4].Recent studies in Cameroon upon the identification of GMO products using PCR based recognition reveals that the 32 products were found to contain genetically modified ingredients [8]. Similar studies in Serbia on imported food products from European Union countries shows none of the products resulted positive in GM testing. This clearly shows, EU made strict regulations upon the GM crops and GM crop based food products [9]. PCR based detection method for the identification of GM in food and feed samples collected from Jordan market ensued 5.2% of positive from 200 food and 80 feed samples [6]. In total 447 food samples containing only soybean products collected during the period of 2006 and 2007 from Serbia markets where analyzed. Even though the investigation exposed the presence of GM products (11%) and most of them are labeled as per regulations, yet mislabeling was found in 10 products which contains the material above the 0.9% [10].
A food should be free from microbial contamination until it reaches the consumers table. All kind of safety measures must be ensured by industries to make it possible. But still microbes are found in food products by any source, leading to the spoilage of food product and causing food borne infections to the consumers. Home cooked food samples collected from the villages in Malawi has shown to be contaminated upto 35% of pathogenic microbes and by further investigation, it was found that contamination of food samples by poor handling and unhygienic conditions [11]. Microbial contamination is a crucial problem in various food industries causing major economic losses. Various industries like dairy, poultry industries follow various cutting-edge technologies to reduce the microbial contamination in end product [12,13]. Microbial contamination can be explored by various physical methods including microscopic and culture depend approaches. But these techniques have various limitations in the confirmation of microbial presence. In present decades, PCR based methods are highly adopted due to its high accuracy and reliability [14].
In the present review, we discuss the utilization of DNA as an effective tool in ensuring the quality and safety in various foods and processed food products (Figure 1).
Figure 1: DNA-An Effective Tool on Food Safety.
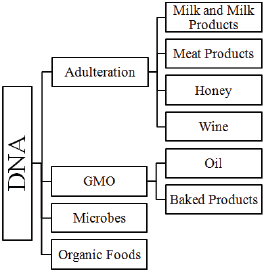
Discussion
Different DNA isolation methods from food products
In general, DNA can be isolated using various standard protocols like Wizard method [15], CTAB protocol [16], also commercial kits are available in the market for the extraction of DNA. These kit's can be utilized to extract DNA from different food samples like, milk and milk products [17], fermented products [18], oils [19], beverages [20]. Apart from the standard protocols and commercial kits, various modified protocols are also adopted by researchers in order to enhance the efficiency of quality and quantity of extracted DNA [21].
DNA based detection of adulteration and GMO in food products
Milk and milk products: Milk stands as a source of high protein based food for vegetarians around the world. Commercial milk is available in the form of cow's milk, buffalo milk, goat milk and sheep milk. The chemical composition of different milk species is almost similar which makes the evaluation of adulterant a high risky process. Since, cow’s milk is cheaper than other milk species; there is a high chance for the addition of cow's milk to other milk species. Similarly, cheese prepared from respective milk has specific characteristics and these adulterations can affect their features, leading to disappointment of the costumer. Therefore, the need of biotechnological approach to recognize the adulteration in every stage of milk processing is important. Aside the adulteration, DNA could be used as an effectual tool in identification of microbial contamination.
A comparative DNA extraction study on ovine milk samples indicates the modified protocols have the greater efficiency over standard protocols and commercial DNA isolation kits [22]. Genomic DNA of 15 cows and 15 goat's milk samples were successfully isolated and using a PCR based technique based on the use of chelex resin. This method can be applied in the identification of specific traits [23]. A similar investigation on adulteration of goat's milk with cow's milk in Taiwan, targeting the specific bovine mitochondrial DNA shown a 25% of adulteration in goat’s milk and 50% of debasement in goat's milk powder [24]. A study with 17 goat cheese and 7 sheep cheese in Czeck Republic markets by targeting the cytochrome b coding sequence in mitochondrial gene pictured 3 goat cheeses and 1 sheep cheese contained undeclared presence of cow’s milk [25]. An evaluation of PCR assay aiming the mitochondrial 12SrRNA gene for the detection of cow's milk in buffalo’s milk in Egyptian markets, resulted 14% of them are purely cow's milk and 38% of them are adulterated with cow’s milk [26]. The DNA diagnostic method targeting mitochondrial 12SrRNA gene was modified and reported as novel method in 2016. The method was investigated with the laboratory adulteration models of milk powders [27].
Wine: One of the most common beverages consumed around the world is wine. In major countries, wine and beer occupy a major portion in their diets. Also, various bioactive compounds present in the wine have been reported for their health benefits like cardiovascular disease prevention, anti-carcinogenic property [28]. Primarily, the quality of wine falls on its vinification process and the grape species. Compounds which are considered as signature, like specific aromatic compounds, monoterpenes, trace compounds are utilized to test the grape varieties [29]. The quality of wine marks its price in a market. Identification of signature compounds from the processed wines cannot provide the exact grape variety utilized as a source. Therefore, it is highly necessary to develop a standard protocol in identification of grape monovariety.
As an attempt, VVMD5, VVMD27, VVMD32, VVMD36 microsatellite loci are used for the identification of monovariety wine species from various cultivators in France. The process of extraction was done on both solid and aqueous fractions. But the results demonstrate that, extraction is more efficient in solid fraction then aqueous part. This may be due to the presence of DNA inhibitors in the aqueous fraction [29]. Late, same microsatellite loci and similar method was used as an attempt to analyze a grape variety Vitis vinifera in commercial must mixtures and also in experimental mixtures. The present study has proved that, DNA can be analyzed from the experimental wines until the last day of the fermentation process [30]. Vitis vinifera nine cis epoxycarotenoid dioxygenase2 (VvNCED2) was used as reference gene in authentication of grape variety in a wine samples. Real time PCR methodology was developed and reported for the DNA presence in aged wines. In addition, the study has quantified Saccharomyces cerevisiae DNA in residual wine samples [21]. Comparable to the previous studies, pure DNA were extracted using an enhanced protocol and quantified using VrZAG79 primer with optimized PCR conditions which can be applicable for both monovarietal and commercial wine samples [31].
Oil: Oil and oil based products like margarine, other edible products act as an active ingredient in day to day life of the people around the world. Every oil species has a respective fatty acid profiles, TAG composition, and phytosterol profiling. To bring down the price of the pure oils and for other reasons, various governments agreed to blend it with other oils at appropriate percentage. But still molecular level authentication is essential to satisfy the consumer.
Initially the researchers tried to extract the soybean DNA from the crude oil and several fractions during processing using Wizard extraction method. The experiment showed a positive result in crude oil samples whereas the method failed to extract the DNA from refined fractions [32]. It shows that, refining process is very well efficient in removal of DNA molecules, from the samples. A subsequent study on extraction of DNA at different stages of refining process showed that, degumming step plays a vital role in removal of DNA [33]. DNA was successfully extracted using CTAB protocol with some modifications in olive oil samples. Further the DNA was taken for AFLP analysis to match the outcomes with the cultivar AFLP results [34]. The major barrier in extraction of genetic material from the oil is its less bioavailability [35]. Late, a sensitive assay using 5s DNA spacers to identify the crop specific food products were developed and utilized to detect the crop specificity in sunflower and maize oil samples obtained commercially [36]. The need for DNA based validation of the oil products got concern to a greater extend and demand towards the optimized which led to various studies towards the derivation of standard methods. A study comparing all the four commercial kit protocols for the DNA extraction from the food sample using different oils such as blended oils, refined oils and oils labeled as GM has shown wizard magnetic method has the higher efficiency of DNA extraction from different oil samples [19].
Honey: Honey, a natural product which is acquired when nectars are collected and stored in honeycombs by bees. For several centuries, honey was utilized as a good source for nutrition and also in medicinal purpose. The properties of honey depend upon their origin. Therefore, it is most important to affiliate the honey with the plant origin. Molecular level detection of plant origin of honey has various advantages then other available methods.
For the first time, DNA in honey samples are isolated to trace, the honey collected from genetically modified Bt (Cry 1 Ac gene) cotton plants which is planted in about 3.7 million hectares during the period of 2004. The extraction and amplification protocols are successful in DNA isolation and identification. A list of six specific primers Sad 1-F, Sad 1-R, 35S1, 35S2, 35 F-S, Bt-R are used in the study to authenticate the Bt gene in the honey samples [37]. As an effort, for validation of protocol for the extraction of DNA from pollen in honey was executed. With a collaboration of 14 labs in and around Germany has accompanied for the establishment of method. Finally, the in-house and interlaboratory validation evidenced the DNA extraction from five different samples (Honeydew Honey with multifloral honey, Wild flower honey "flowers of the mountain", Wild flower honey, Rape honey, Acacia-with multifloral honey) [38]. With the comparison of previous studies, an improved method for the efficient DNA extraction from honey samples were performed using commercial DNA isolation kits, finally stating that Wizard method with pretreatment has the maximum yield in the aspect of both purity and quantity [39].
Baked products: Baked food stuffs consisting of biscuits, cake, waffle, etc., are mostly consumed by the school children. Soy based ingredients are the widely used for the production of these products. In order to ensure the safety regarding the addition of GM Soy products in food process, standard protocols must be developed in detection of GM adulteration.
Variations in buffer volumes and sample sizes provided a good amplification results using PCR technique targeting soy lectin gene in analysis of GMO adulteration of soy based products such as chocolate and biscuits. In comparison of 5 different DNA isolation techniques, CTAB and Nucleon PhytoPure Kit yielded good results for chocolate products, whereas in case of biscuits CTAB and Genespin DNA Isolation Kit gave best results [40].
Meat Products: Meat production and consumption has tripled in last 3 to 4 decades. Pork, poultry, beef and mutton are mostly consumed around the globe. Yet, the fraudulent in the meat products could not recognized by naked eye. A distinct technique is continuously required to guarantee the costumer safety.
At first, differences in the 18s rRNA gene was used to differentiate between the meat products [41]. Later researchers have found the advantages on single primer over multi primer in detection of target gene in the meat product authentication. Act in gene associated PCR test provided an effective outcome on chicken and turkey identification in meat admixtures [42]. Differentiation of chicken in turkey meat and vice versa could be achieved by the study on variability in intron actin gene locus [43]. A PCR based protocol was estimated in order to detect the presence of pork in heated and nonheated ground beef and pate products. Contamination was found upto 1% in the meat products while carrying out 20 PCR cycles results satisfied quantity of amplified products than25 and 30 PCR cycles [44]. However, 30 cycles species specific real time (TaqMan) PCR technique targeting the mitochondrial cytochrome b (cytb) gene to detect beef, pork, lamb, chicken and turkey species even at the 0.5% admixture [45].
Organic foods
Consumers believe that organic foods are safer and healthier than conventional foods. Generally, organic foods are sold at premium price in the markets [46]. Nitrogen isotope signature method has been reported as an effective technique in differentiating the organic foods from non-organic foods [47].
Here we review the application of DNA to differentiate the organic foods from non-organic food. Pesticides applied on the plant are mostly a known carcinogenic agent. These carcinogenic compounds can bound to the segment of plant DNA and may generate DNA adducts. A study on radio chromatograms using 32P post labeling in the plants such as grapes, bush beans, soybeans, pumpkins, and cucumbers treated with pesticides has shown high level of DNA adducts despite of untreated plants. The study also indicates that treated plants have undergone severe oxidative stress and lipid per oxidation [48]. Thus, DNA adducts could be used as biomarker in differentiation of organic foods from non- organic foods.
Microbes
Food and processed food products should be monitored at regular intervals to control the unwanted microbial load in the final product. Standard microbial count estimation techniques are time consuming process. Rapid techniques are needed to overcome the standard techniques with added accuracy in specificity.
PCR-DGGE (Denaturing gradient gel electrophoresis) was one of the recent methods used to detect the microbial community from various food products such as water, beverage, dairy and fermented products. PCR-DGGE technique involves simple steps like extraction of microbial genetic material followed by PCR amplification of variable regions of ribosomal DNA and finally DGGE analysis for species identification [14].
Conclusion
Our review on using DNA as an effective tool on food safety on various aspects such as detection of adulteration, GMO content, microbial load and also differentiation of organic foods from non- organic foods supports the supposition to use this technique for food safety. Since, countless embodiments of fraud in food product continue to evolve; advanced method on detection may provid great accuracy over previous methodologies. However, regulatory bodies around the globe should provide severe regulations on food adulteration in-order to afford consumer safety.
References
- Travers A, Muskhelishvili G (2015) DNA structure and function. FEBS Journal 282(12): 2279-2295.
- Zhang C, Wohlhueter R, Zhang H (2016) Genetically modified foods: A critical review of their promise and problems. Food Science and Human Wellness 5(3): 116-123.
- Buiatti M, Christou P, Pastore G (2013) The application of GMOs in agriculture and in food production for a better nutrition: Two different scientific points of view. Genes Nutr 8(3): 255-270.>
- Abdel-Mawgood AL, Gassem MA, Alsadon AA, Alghamdi SS, Al-Doss AA (2010) Monitoring of genetically modified food in Saudi Arabia. African Journal of Food Science 4(8): 536-540.
- Salameen AF, Kumar V, Aqeel AH, Hashash AH, Hejji BA (2012) Detection of genetically modified DNA in fresh and processed foods sold in Kuwait. GM Crops Food 3(4): 283-288.
- Herzallah SM (2012) Detection of genetically modified material in feed and foodstuffs containing soy and maize in Jordan. Journal of Food Composition and Analysis 26(1-2): 169-172.
- Nikolic Z, Taski-Ajdukovic K, Tatic M, Balesevic-Tubic S (2009) Monitoring of the roundup ready soybean in the vojvodina province in Serbia. Industrial Crops and Products 29(2-3): 638-641.
- Roger DD, Gone S (2014) Evidence of the presence of genetically modified foods in the sudano-sahelian zones of cameroon. Food and Nutrition Sciences 5(10): 922-928.
- Zdjelar G, Nikolic Z, Vasiljevic I, Bajic B, Jovicic D, et al. (2013) Detection of genetically modified soya, maize, and rice in vegetarian and healthy food products in Serbia. Czech Journal of Food Sciences 31(1): 43-48.
- Nikoli Z, Ta K, Ajdukovi K, Jevti A, Marinkovi D (2009) Detection of GM soybean in food products by simultaneous employment of three pairs of PCR primers. Food Research International 42(3): 349-352.
- Taulo S, Wetlesen A, Abrahamsen R, Kululanga G, Mkakosya R (2008) Microbiological hazard identification and exposure assessment of food prepared and served in rural households of Lungwena, Malawi. International Journal of Food Microbiology, 125(2): 111-116.
- Walia K, Lynch H, Argüello H, Grant J, Leonard FC, et al. (2016) The efficacy of different cleaning and disinfection procedures to reduce Salmonella and Enterobacteriaceae in the lairage environment of a pig abattoir. Int J Food Microbiol 246: 64-71.
- Alexopoulos A, Plessas S, Kourkoutas Y, Stefanis C, Vavias S, et al. (2017) Experimental effect of ozone upon the microbial flora of commercially produced dairy fermented products. International Journal of Food Microbiology 246: 5-11.
- Ercolini D (2004) PCR-DGGE fingerprinting: Novel strategies for detection of microbes in food. J Microbiol Methods 56(3): 297-314.
- Hemmer W (1997) Foods derived from genetically modified organisms and detection methods. Report 2/97, Basel, Switzerland, pp. 14-20.
- Jankiewicz A, Broll H, Zagon J (1999) The official method for the detection of genetically modified soybeans (German Food Act LMBG § 35): a semi-quantitative study of sensitivity limits with glyphosate- tolerant soybeans (Roundup Ready) and insect-resistant Bt maize (Maximizer). European Food Research and Technology 209(2): 77-82.
- Volk h, Piskernik S, Kurinčič M, Klančnik A, Toplak N, et al. (2014) Evaluation of different methods for DNA extraction from milk. Journal of Food & Nutrition Research 53(2): 97-105.
- Djurkin Kušec I, Radišić Ž, Komlenić M, Kusec G (2015) Comparison of commercial DNA kits and traditional DNA extraction procedure in PCR detection of pork in dry/fermented sausages. Poljoprivreda 21(1): 199202.
- Costa J, Mafra I, Amaral JS, Oliveira MBPP (2010) Detection of genetically modified soybean DNA in refined vegetable oils. European Food Research and Technology 230(6): 915-923.
- Martins-Lopes PF, Pereira L, Guedes-Pinto H (2011) Method and Kit for DNA extraction from Vitis vinifera L and for Amplification and detection of Grapevine Varieties or Cultivars in Musts or Wines.
- Savazzini F, Martinelli L (2006) DNA analysis in wines: Development of methods for enhanced extraction and real-time polymerase chain reaction quantification. Analytica Chimica Acta 563(1-2): 274-282.
- Psifidi A, Dovas CI, Banos G (2010) A comparison of six methods for genomic DNA extraction suitable for PCR-based genotyping applications using ovine milk samples. Molecular and Cellular Probes 24(2): 93-98.
- Amills M, Francino O, Jansa M, Sanchez A (1997) Isolation of genomic DNA from milk samples by using Chelex resin. The Journal of Dairy Research 64(2): 231-238.
- Cheng YH, Der Chen S, Weng CF (2006) Investigation of goats' milk adulteration with cows' milk by PCR. Asian-Australasian Journal of Animal Sciences 19(10): 1503-1507.
- Maskova, E, Pauli'ckova I (2006) PCR-based detection of cow's milk in goat and sheep cheeses marketed in the Czech Republic. Czech Journal of Food Sciences, 24(3): 127-132.
- Darwish SF, Allam A, Amin AS (2009) Evaluation of PCR Assay for Detection of Cow a€TM s Milk in Water Buffalo a€TM s Milk World 7(4): 461-467.
- Liao J, Liu YF, Ku T, Liu MH, Huang Y (2017) Qualitative and quantitative identification of adulteration of milk powder using DNA extracted with a novel method. Journal of Dairy Science 100(3): 1657-1663.
- Guerrero RFG (2009) Wine, resveratrol and health: a review. Natural Product Communications 4(5): 635-658.
- Siret R, Boursiquot, Merle MH, Cabanis JC, This P (2000) Toward the authentication of varietal wines by the analysis of grape (Vitis vinifera L.) Residual DNA in must and wine using microsatellite markers. Journal of Agricultural and Food Chemistry 48(10): 5035-5040.
- Siret R, Gigaud O, Rosec JP, This P (2002) Analysis of grape Vitis vinifera L. DNA in must mixtures and experimental mixed wines using microsatellite markers. Journal of Agricultural and Food Chemistry 50(13): 3822-3827.
- Pereira L, Guedes PH, Martins PL (2011) An enhanced method for vitis vinifera L. DNA extraction from wines. American Journal of Enology and Viticulture 62(4): 547-552.
- Pauli U, Liniger M, Zimmermann A (1998) Detection of DNA in soybean. European Food Research & Technology 207(4): 264-267.
- Gryson N, Ronsse F, Messens K, De Loose M, Verleyen T (2002) Detection of DNA during the refining of soybean oil. J Am Oil Chem Soc 79(2): 171174.
- Busconi M, Foroni C, Corradi M, Bongiorni C, Cattapan F, et al. (2003) DNA extraction from olive oil and its use in the identification of the production cultivar. Food Chemistry 83(1): 127-134.
- Gryson N, Messens K, Dewettinck K (2004) Influence of different oil- refining parameters and sampling size on the detection of genetically modified DNA in soybean oil. J Am Oil Chem Soc 81(3): 231-234.
- Doveri S, Lee D (2007) Development of sensitive crop-specific polymerase chain reaction assays using 5S DNA: Applications in food traceability. Journal of Agricultural and Food Chemistry 55(12): 46404644.
- Cheng H, Jin W, Wu H, Wang F, You C, et al. (2007) Isolation and PCR detection of foreign DNA sequences in bee honey raised on genetically modified Bt (Cry1Ac) cotton. Food and Bioproducts Processing 85(C2): 141-145.
- Waiblinger HU, Ohmenhaeuser M, Meissner S, Schillinger M, Pietsch K, et al. (2012) In-house and interlaboratory validation of a method for the extraction of DNA from pollen in honey. Journal Fur Verbraucherschutz Und Lebensmittelsicherheit 7(3): 243-254.
- Soares S, Amaral JS, Oliveira MBPP, Mafra I (2015) Improving DNA isolation from honey for the botanical origin identification. Food Control 48: 130-136.
- Gryson N, Messens K, Dewettinck K (2004) Evaluation and optimisation of five different extraction methods for soy DNA in chocolate and biscuits. Extraction of DNA as a first step in GMO analysis. Journal of the Science of Food and Agriculture 84(11): 1357-1363.
- Matsunaga T, Shibata K, Yamada J, Shinmura Y, Chikuni K (1998) Identification of meat species based on the difference of 18S ribosomal RNA Genes. Nippon Shokuhin Kagaku Kogaku Kaishi 45(12): 719-723.
- Hopwood AJ, Fairbrother KS, Lockley AK, Bardsley RG (1999) An actin gene-related polymerase chain reaction (PCR) test for identification of chicken in meat mixtures. Meat Science 53(4): 227-231.
- Lockley Ak, Bardsley RG (2002) Intron variability in an actin gene can be used to discriminate between chicken and turkey DNA. Meat Science 61(2): 163-168.
- Calvo JH, Osta R, Zaragoza P (2002) Quantitative PCR detection of pork in raw and heated ground beef and pate. Journal of Agricultural and Food Chemistry 50(19): 5265-5267.
- Doole JJ, Paine KE, Garrett SD, Brown HM (2004) Detection of meat species using TaqMan real-time PCR assays. Meat Sci 68(3): 431-438.
- Capuano E, Boerrigter ER, Vander VG, Van Ruth SM (2013) Analytical authentication of organic products: An overview of markers. Journal of the Science of Food and Agriculture 93(1): 12-28.
- Sturm M, Lojen S (2011) Nitrogen isotopic signature of vegetables from the Slovenian market and its suitability as an indicator of organic production. Isotopes in Environmental and Health Studies 47(2): 214220.
- Boerth DW, Eder E, Stanks JR, Wanek P, Wacker M, et al. (2008) DNA adducts as biomarkers for oxidative and genotoxic stress from pesticides in crop plants. Journal of Agricultural and Food Chemistry 56(15): 67516760.
© 2017 Duraimurugan K, et al. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






