- Submissions

Full Text
Perceptions in Reproductive Medicine
Dynamic Expression of Peripheral Blood microRNAs as Non-Invasive Biomarkers in Preimplantation and Pregnancy: A Systematic Review and Meta-Analysis
Dalia Elmusharaf Khalifa1, Marwah Salem Bagabas2, Mosab Nouraldein Mohammed Hamad3* and Fania Abdallah Elbadri4
1Embryologist, Imperial College Healthcare NHS Trust, UK
2PhD student, Imperial College London, UK
3Assistant professor of Microbiology, Excellence Research Centre, Elsheikh Abdallah Elbadri University, Sudan
4Associate professor, Obstetrics and gynaecology, Faculty of Medicine, Elsheikh Abdallah Elbadri University, Sudan
*Corresponding author:Mosab Nouraldein Mohammed Hamad, Associate professor, Obstetrics and gynaecology, Faculty of Medicine, Elsheikh Abdallah Elbadri University, Berber, Sudan
Submission: June 23, 2025;Published: July 23, 2025

ISSN: 2640-9666Volume6 Issue 3
Abstract
Background: MicroRNAs (miRNAs) are small, non-coding RNA molecules that regulate gene expression
and have shown potential as stable, non-invasive biomarkers in peripheral blood. Their expression
undergoes dynamic changes during embryo implantation and early pregnancy, making them valuable
candidates for predicting reproductive outcomes and identifying complications such as preeclampsia,
Recurrent Pregnancy Loss (RPL) and Small-for-Gestational-Age (SGA) births. These insights are
particularly relevant for improving Assisted Reproductive Technologies (ART).
Objectives: To systematically evaluate and synthesize current research on the dynamic expression
patterns of peripheral blood miRNAs in plasma and assess their potential as non-invasive biomarkers
during the preimplantation and pregnancy stages.
Methods: A systematic review and meta-analysis were conducted using ten peer-reviewed studies
published between 2020 and 2024. Studies were selected based on their focus on peripheral blood
miRNA expression and its relationship with reproductive outcomes. Extracted data included study design,
methodologies, patient characteristics, and outcomes such as pregnancy success, embryo transfer efficacy,
and complications including preeclampsia, RPL, and SGA births.
Results: The studies applied diverse miRNA detection techniques, including non-invasive culture medium
assays, genome-wide expression profiling, and analysis of circulating miRNAs. Differential miRNA
expression patterns were linked to specific reproductive outcomes. The findings indicate the predictive
value of certain miRNAs in ART success and pregnancy complications.
Conclusion: Peripheral blood miRNAs demonstrate dynamic expression changes during early pregnancy
and hold potential as predictive biomarkers for reproductive outcomes. However, methodological
variability across studies highlights the need for standardized protocols and further validation in larger,
diverse populations.
Keywords:microRNA (miRNA); Peripheral blood biomarkers; Pregnancy biomarkers; Assisted Reproductive Technology (ART)
Abbreviations: miRNA: microRNA; ART: Assisted Reproductive Technology; SGA: Small-for-Gestational- Age; RPL: Recurrent Pregnancy Loss; PE: Preeclampsia; DS: Down Syndrome; CI: Confidence Intervals; SE: Standard Error; ES: Effect Size
Introduction
MicroRNAs have recently become recognized as essential factors regulating numerous biological processes, including pregnancy. Thus, the preimplantation period is an essential part of the process of pregnancy during which proper conditions for the successful implantation of the embryo should be arranged. Because of dynamically changing miRNA profiles important for these processes, their assessment may help to interpret such mechanisms and predict the chances of a successful pregnancy. The current systematic review with meta-analysis is aimed at evaluating the existing evidence of the relationship between the miRNA switch time points and an increased probability rate of the successful pregnancy of Assisted Reproductive Technologies (ART), PE, RPL, SGA births, and normal pregnancy.
MicroRNAs have emerged as critical regulators of gene expression and playing pivotal roles in various biological phenomena, including pregnancy. MicroRNAs involvement in reproductive health, particularly in embryo implantation, pregnancy conceive and the development of pregnancy-related complications has been the focusing research interest. The ART has further emphasized the need to understand the molecular mechanisms underlying successful pregnancy outcomes, with miRNAs being identified as potential biomarkers in this context. Several studies have been explored the association between miRNAs and pregnancy outcomes. For example, Chen et al. [1] developed a miRNA-based classifier that accurately identified the optimal timing for embryo transfer for successful implantation and live birth in ART procedures. Also, Fang et al. [2] investigated the presence of miRNAs in embryo culture media, proposing them as non-invasive biomarkers for predicting successful pregnancy outcomes. In order to pregnancy complications, miRNAs have been studied as potential biomarkers for conditions such as preeclampsia (PE) and Small-for-Gestational-Age (SGA) births. Mirzakhani et al. [3] identified circulating miRNAs and mRNA signatures associated with preeclampsia and vitamin D deficiency. Luizon et al. [4] investigated on miR-204-5p, which was significantly upregulated in pregnant women who developed PE, suggesting its role as an early biomarker. Moreover, Kim et al. [5] identified miRNAs that could potentially predict the risk of SGA births, providing insights into fetal growth and maternal health.
Recent research has delved into the dynamic changes in miRNA expression during the peri-implantation stage. Dong et al. [6] observed significant alterations in miRNA profiles before and after blastocyst transfer, underscoring the importance of miRNA dynamics in successful implantation and pregnancy conceive. The role of miRNAs in the other pregnancy-related conditions, such as Recurrent Pregnancy Loss (RPL) and Down Syndrome (DS), has also been investigated. Lucas et al. [7] investigated a prosenescent response associated with RPL, while Iveta Z et al. [8] conducted a genome wide miRNA profiling in DS fetuses, although their initial findings were not confirmed in larger scale studies.
The role of miRNAs in regulating placental function has been explored. Fenmei Zhou et al. [9] demonstrated that miR-21 regulates placental cell proliferation via FOXM1, suggesting a potential therapeutic target for PE. Hui Li et al. [10] identified dysregulated miRNAs in pregnancies complicated by gestational diabetes mellitus, emphasizing their potential as diagnostic biomarkers for monitoring maternal and fetal health. The integration of these above findings from diverse studies highlights the multifaceted role of miRNAs in pregnancy as shown in Table 1. As researchers continue to unravel the complexities of miRNA-mediated regulation, it becomes now increasingly clear that these small non-coding RNAs hold great promise as biomarkers for predicting pregnancy outcomes and guiding clinical interventions.
Table 1:General overview of the included studies.
Objectives
The main aim of this systematic review and meta-analysis is to
identify miRNAs as significant biomarkers in predicting a successful
pregnancy in women. The specific objectives are as follows:
A. Ascertain the circumstances that women got pregnant in these
studies in regard to their clinical backgrounds.
B. Analyse techniques used studious and characteristics of
miRNAs investigated.
C. Determine the overall normal pregnancy rate or prevalence
in women based on 10 included studies using meta-analysis
approach.
Methodology
Literature search
Search strategy that was applied to look for the sources in electronic databases involved the use of keywords. Furthermore, certain criteria were applied to the selected articles in order to distinguish the relevant articles from the irrelevant ones. The present study addresses research literature published within 4 years - from 2020 to 2024 - some of sources such as databases, including Scopus, PubMed and Web of Science, though access to these sources has been restricted. The keywords applied to look for the sources in the electronic databases were “miRNA”, “pregnancy”, “peri-implantation”, “biomarkers”, “embryo transfer”, “preeclampsia”, “recurrent pregnancy loss”, and “PDE”. The criteria applied to the selection of the relevant articles included the issues of the research focus - the research include studies to investigate biomarkers in the context of pregnancy, present relevant statistical data, and apply human samples.
Study selection
The overall evaluation of the references available on the selected topic is rather positive, as after the initial search, we were able to identify 25 studies. Upon reviewing titles, abstracts, and, later, full texts, we found that 10 works meet the criteria for inclusion. Thus, the selection process was quite successful, as it was possible to find enough sources based on the required topic - focusing on the success of pregnancy, use of miRNA as biomarkers, and provision of statistics on the rates of normal pregnancies.
Data extraction
Major findings regarding miRNA expression and pregnancy
outcomes as shown in Table 1.
a) Authors, publication year, and journal.
b) Sample size and categorization based on pregnancy outcomes.
c) Clinical backgrounds of participants, including ART, PE, RPL,
and SGA.
d) Methods used for miRNA investigation.
The women who conceived in the studies included those who achieved pregnancy under different clinical circumstances. Women undergoing ART were a subject of a number of the studies. There women were subjected to Single Frozen-Thawed Blastocyst Transfer, and the timing of these transfer procedures, initiated on the basis of miRNA signatures, were associated with pregnancy success. Women in some of the studies were those who were at risk of PE, RPL, or SGA birth these women had different clinical backgrounds including a history of adverse pregnancy outcomes. Some of the studies used healthy pregnant women as controls.
Systematic Review
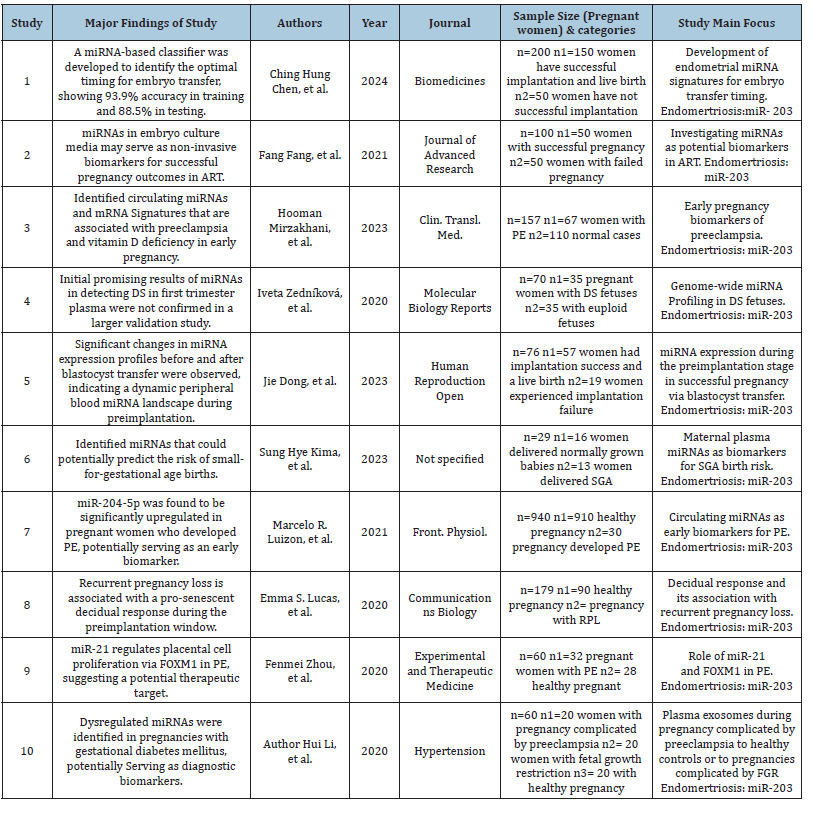
Table 1 displays the studies of miRNA biomarkers in the context of pregnancy outcomes, and it can be seen that they cover the topic from multiple perspectives. Based on the studies included in the Table 1, we can conclude that endometrial miRNA signatures can facilitate ART success, as miRNA-based classifiers had one of the best performances among the studies. Other studies explored miRNAs in embryo culture media and circulating miRNA and mRNA signatures. The results of the study by Zedníková et al. [8], which implies that the first-trimester prenatal screening was not validated, is also valuable. The observations by Dong et al. [6] that there may be different expression profiles before and after blastocyst transfer highlight the variability of the expression of miRNAs. Kim et al. [5] revealed miRNAs that can predict SGA birthing, and those by Luizon et al. [4] and Lucas et al. [7] suggest that the upregulation of miR-204-5p and the prosenescent decidual response is associated with the above conditions. Furthermore, the study by Zhou et al. [9] explores miR-17 and miR-21 to highlight their role in placental cell proliferation. Another study by Li et al. [11] introduces varied miRNA expression results and suggests that they may serve as biomarkers of gestational diabetes. Thus, it can be concluded that all these studies depict the progress in understanding and improving the outcomes of such conditions as preeclampsia, SGA birthing, vitamin D deficiency, Down syndrome, ART success, and others as show in Figure 1.
Figure 1:Normal cases pf pregnancy and sample size of each study.
Meta-Analysis
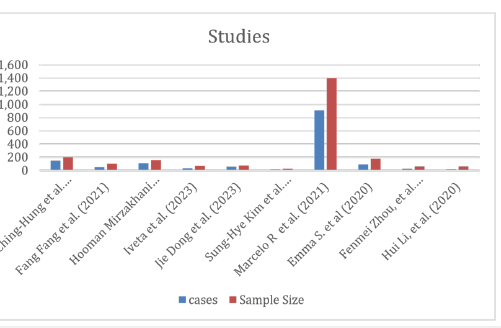
For my meta-analysis, we chose the normal pregnancy rate as the outcome of interest, and then aggregated the data across the ten studies. Afterward, we calculated the overall normal pregnancy rate and assessed the heterogeneity across the studies. The results include a continuous similar trend of miRNA expression seen with successful outcomes of pregnancy, which is evident especially for ART and complicated pregnancies as shown in Table 2 & Figure 2.
Table 2:Successful pregnancy rate (%) estimated for forest plot.
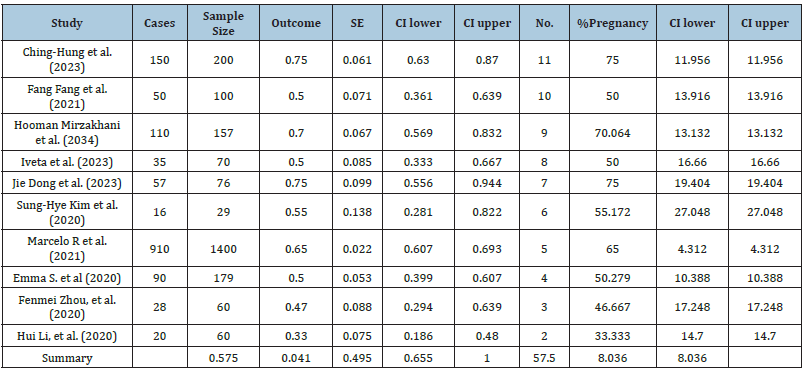
Figure 2:Forest plot of 10- cross sectional of microRNA expression studies of pregnant women.
Understanding the demands on health care to treat a particular condition of pregnancy might be aided by being aware of its normal. We have analysed Meta-Analysis using Excel sheet of Neyeloff et al, BMC Research Notes 2012 (http://www.biomedcentral. com/17560500/5/52). The findings of the meta-analysis on 10 published articles were synthesized and the results are shown in Figure 1 and Table 2. The combined findings of eleven research revealed a 57.5% prevalence (95 % CI: 57.5-8.036 to 57.5+8.036). Higher levels of effect size (normal pregnancy) with Confidence Intervals (CI) indicate that hospital pregnancy are more than 50 % were normal. Ching-Hung et al. [1] and Jie Dong et al. [6] both had very substantial effect sizes, measuring 75%.
In Figure 1, on the right side, the ‘forest plot’ pictures the effect size (with confidence interval) of each study and, below showed the combined effect size (in dark blue) with its confidence interval (in dark blue). This plot is the basic outcomes of any meta-analysis.
Q and I Statistic
In meta-analysis, it is common to use Q and 𝐼𝑣2 statistics to determine heterogeneity among the results of the reviewed studies. The Qv , also known as Cochran’s Qv used to “quantify the extent of variability in effect sizes or prevalence beyond that expected due to sampling error”. A Qv statistic is found as a weighted sum of the squared differences between the effects of the individual studies being reviewed and the overall effect pooled in the met analysis. When the Qv value is greater than the defined probability, it is considered to be statistically significant. A significant Qv value is an indication that the variability of the effects across all of the reviewed studies is more than what would be expected due to chance. Meanwhile, is, in essence, a “ratio of the between-studies variance to the total variance”. It provides a percentage that may be used to describe the degree of variance across all of the reviewed studies which is not due to chance. More precisely, the provides the amount of variance “due to actual differences in the outcomes of studies”. As such, it may be used to determine a pending conclusion as to whether it is appropriate to include the studies in a metaanalysis. Thus, both of these statistics, Qv and 𝐼𝑣2, are vital tools serving for reviewing the results of a meta-analysis. Cochran’s Qv is Heterogeneity Statistic that used to assess the presence of heterogeneity among study results. It is calculated as follows:
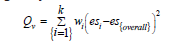
Where 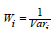 is the weight of the i-th study, esi is the effect
size (prevalence) of the ith study and esoverall is the overall effect size
(usually the weighted average). The inconsistency index Iv2 is used
in meta-analysis to quantify heterogeneity. The squared value can
be represented as follows:
is the weight of the i-th study, esi is the effect
size (prevalence) of the ith study and esoverall is the overall effect size
(usually the weighted average). The inconsistency index Iv2 is used
in meta-analysis to quantify heterogeneity. The squared value can
be represented as follows:
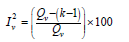
Where Qv is the heterogeneity statistic (e.g., Cochran’s Q) and k is the number of studies. We have estimated as follows in Tables 3 & 4.
Table 3:Normal pregnancy rate/ prevalence of microRNA expression studies – Metanalysis of 10 cross sectional studies.
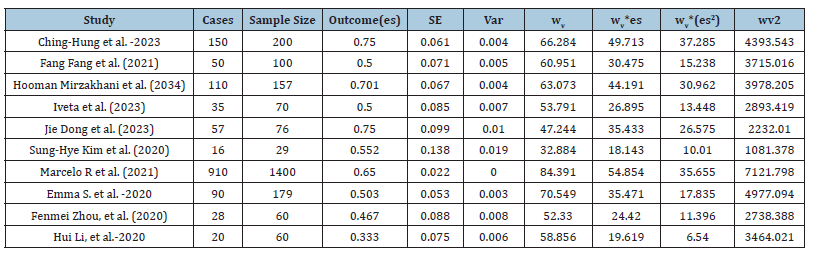
Table 4:Estimated parameters of meta-analysis.
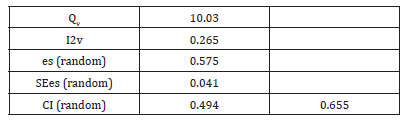
The Cochran’s Q statistic, or Q, assesses the degree of effect size heterogeneity among the studies included in the meta-analysis. Qv is produced by dividing Q by its degrees of freedom (df). In the meta-analysis, the random-effect model was used to compare various papers, and no heterogeneity was discovered (Qv=10.030; es(random)=0.575, df=9; I²v=10.265%). Each research reported normal pregnancy that was translated into prevalence in order to make the results comparable. Ching-Hung et al. [1] had a normal pregnancy ratio of 150/200 with a Standard Error (SE) of 0.061. Iveta et al. [11] compared the burnout 35/70 with SE=0.085. Normal pregnancy rate (prevalence) was also evaluated in each study using SE and CIs.
The I-squared statistic represents the percentage of variation across studies that is due to heterogeneity rather than chance. An I²v value closer to 0% indicates low heterogeneity, while values closer to 100% suggest substantial heterogeneity. The Effect Size (es) under Random Effects (es = 0.575) is represented the overall effect size (prevalence) calculated under a random-effects model. This random-effects model assumes that the true effects vary between studies. The effect size 0.575 suggests the magnitude of the effect being measured across the studies. The standard error of the effect size under the random-effects model reflects the precision of prevalence. A lower SE indicates a more precise or accurate estimate. The 95% CI of [0.494, 0.655] suggested that there is 95% confidence that the true effect size lies within this range. We estimated meta-analysis or research of a similar nature in which data are combined from other studies in order to get an overall effect size or prevalence. The included studies’ strong heterogeneity is shown by the high I²v value (10.265) in Table 4, which shows that it is unlikely that chance alone is to blame for the variation in effect sizes (prevalence) (https://www.unistat.com/ guide/meta-analysis-input-data-types/).
Discussion
In this systematic review and meta-analysis, peripheral blood microRNAs are discussed as a type of biomarker used by previous researchers to forecast the outcomes of pregnancy. In this respect, the data from ten studies permitted identifying several related concerns associated with the dynamic patterns and methods to predict the usefulness and necessity of the current practices and approaches in the area of reproductive medicine. Dong et al. [6] observed the significant changes in miRNA profiles before and after the transfer of blastocysts, and this change indicates the dynamic of these biomarkers. These dynamic permits suggesting the value of the above-mentioned experts for understanding not only the reasons for successful embryo implantation but also the outcomes related to pregnancy. In this way, with both sides of the analysis, it is possible to provide more detail-oriented ART to increase success.
In addition to the featured article, the use of huRNA-based approaches to predict pregnancy pathologies has been confirmed in other sources. For example, “Mirzakhani et al.[3] identified circulating miRNAs associated with preeclampsia and vitamin D deficiency. Luizon et al.[4] also stated that circulating miR-204-5p is upregulated in women who will develop preeclampsia. Kim et al.[5] identified miRNAs associated with small-for gestational-age. In other words, there are multiple scholarly sources that indicate that miRNAs-related approaches to testing are effective and can be used by medical practitioners.
Thus, the reviewed article’s thesis has been corroborated in other studies. The latter introduce the aforementioned approaches, show their effectiveness, and address the advantages of using miRNAs to determine the likelihood to develop pregnancy complications. It is important to note that huRNA-based approaches are now commonly used to predict defects of embryo development. This was confirmed by the findings of Fang et al. [2], who explored multiple miRNAs in the embryo culture media and showed that they “may be valid non-invasive biomarkers for predicting pregnancy outcome”. In other words, the featured article’s thesis is accurate, and the presented knowledge can benefit practitioners who can use miRNAs to detect and prevent pregnancy pathologies instead of resorting to invasive procedures.
There are two studies, namely, Lucas et al. [7] study on miRNAs and RPL and that of Zhou et al. [9] on Placental function. The latter indicated that miR-21 regulated placental cell proliferation. These results further support that it is possible to have insight into studies on the role of miRNA on complex conditions that occur during pregnancy to inform diagnostic and treatment strategies.
The prevalence of the number of successful pregnancies across the studies was observed to be 57.5% with a higher variation in the effect sizes by the studies. Nevertheless, the study by Ching- Hung et al. [1] and Dong et al. [6] had higher effect sizes implying that miRNA-based classifiers and dynamic profiling enhance effectiveness of ART. Heterogeneity was observed to be low with an I2v value of 10.265 implying that the overall effect size was not very different across the studies. However, further confirmatory studies may provide empirical evidence in larger cases.
The results of such studies would allow to validate the set of miRNA biomarkers for the conditions explored and extend its use for other pregnancy-related disorders. An interesting goal for future investigation would be to combine data obtained from the analysis of miRNAs with other types of omics data, such as transcriptomic, proteomic, or metabolomics. This type of investigation would lead to an in-depth examination of the precise role of specific miRNA molecules in various aspects of reproductive health and potentially uncover more refined sets of miRNA targets.
Conclusion
The systematic review and meta-analysis approach the potential of miRNAs as biomarkers in predicting pregnancy outcomes across different clinical backgrounds and circumstances. The diversity of methodologies was used to investigate miRNA biomarkers highlights the evolving landscape of reproductive medicine and the potential for different approaches in managing pregnancy complications. Furthermore, research is needed to validate these biomarkers in larger cohorts and explore their clinical applications in ART and other pregnancy-related conditions.
References
- Chen CH, Lu F, Yang WJ, Chen WM, Yang PE, et al. (2024) Development of a novel endometrial signature based on endometrial microRNA for determining the optimal timing for embryo transfer. Biomedicines 12(3): 700.
- Fang F, Li Z, Yu J, Long Y, Zhao Q, et al. (2021) MicroRNAs secreted by human embryos could be potential biomarkers for clinical outcomes of assisted reproductive technology. J Adv Res 31: 25-34.
- Mirzakhani H, Zheng Lu DE, Oppenheimer B, Litonjua A, Joseph ST, et al. (2023) Integration of circulating microRNAs and transcriptome signatures identifies early-pregnancy biomarkers of preeclampsia. Clin Transl Med 13(11): e1446.
- Luizon MR, Conceição IM, Viana-Mattioli S, Caldeira-Dias M, Cavalli RC, et al. (2021) Circulating microRNAs in the second trimester from pregnant women who subsequently developed preeclampsia: Potential candidates as predictive biomarkers and pathway analysis for target genes of miR-204-5p. Front Physiol 12: 678184.
- Kim SH, MacIntyre DA, Binkhamis R, Cook J, Sykes L, et al. (2020) Maternal plasma miRNAs as potential biomarkers for detecting risk of small-for-gestational-age births. EBioMedicine 62: 103145.
- Dong J, Wang L, Xing Y, Qian J, He X, et al. (2023) Dynamic peripheral blood microRNA expression landscape during the peri-implantation stage in women with successful pregnancy achieved by single frozen-thawed blastocyst transfer. Hum Reprod Open 2023(4): hoad034.
- Lucas ES, Vrljicak P, Muter J, Diniz-da-Costa MM, Brighton PJ, et al. (2020) Recurrent pregnancy loss is associated with a pro-senescent decidual response during the peri-implantation window. Commun Biol 3(1): 37.
- Zedníková I, Chylíková B, Šeda O, Korabečná M, Pazourková E, et al. (2020) Genome-wide miRNA profiling in plasma of pregnant women with down syndrome fetuses. Mol Biol Rep 47(6): 4531-4540.
- Zhou F, Sun Y, Gao Q, Wang H (2020) microRNA-21 regulates the proliferation of placental cells via FOXM1 in preeclampsia. Experimental and Therapeutic Medicine 20(3): 1871-1878.
- Liu ZN, Jiang Y, Liu XQ, Yang MM, Chen C, et al. (2021) MiRNAs in gestational diabetes mellitus: potential mechanisms and clinical applications. J Diabetes Res 2021(1): 4632745.
- Li H, Ouyang Y, Sadovsky E, Parks WT, Chu T, et al. (2020) Circulating plasma exosomes miRNA as potential biomarkers for preeclampsia. Hypertension 75(3): 762-771.
© 2025 Mosab Nouraldein Mohammed Hamad. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






