- Submissions

Full Text
Novel Approaches in Cancer Study
Circulating DNA in Serum as a Tumor Marker in Precancerous Lesions of the Cervix in Mexican Women
Zuñiga-Martínez Lourdes1, Porras-Flores Olimpia2, Terán-Figueroa Yolanda3, García- Guerrero Israel4 and Alcántara-Quintana Luz Eugenia1*
1Coordinación para la Innovación y Aplicación de la Ciencia y la Tecnología (CIACyT), México
2Clínica de Displasias, México
3Facultad de Enfermería y Nutrición, México
4Hospital Central, Mexico
*Corresponding author:Coordinación para la Innovación y Aplicación de la Ciencia y la Tecnología (CIACyT), San Luis Potosí, SLP, México
Submission: June 16, 2021 Published: July 06, 2021

ISSN:2637-773XVolume6 Issue1
Abstract
Introduction: Cervical cancer is the fourth leading cause of death in the world. This poor prognosis is due to the fact that in two out of three cases the disease is diagnosed in advanced stages. Consequently, advances in detection methods can lead to early diagnosis of the disease and consequently to an improved prognosis. Methods based on serum-based tumor markers are a promising tool for early diagnosis and in general for biochemical screening of cancer patients. They also offer the added value of being non-invasive methods. Evidence has been found for the release of cellular DNA into the blood, so its quantification has the potential to serve as a tumor marker. If this DNA is detected in serum, it can reflect tumor size, viral load and response to treatment.
Objective: To determine the amount of circulating DNA in serum of patients with precancerous lesions.
Methods: The study was carried out with 40 healthy women and 120 women with precancerous lesions, who attended gynecological-obstetric care. Of the 120, 60 had low-grade lesions and 60 had high-grade lesions. All participants had a blood sample taken for determination of circulating DNA in serum. Specific oligonucleotides were used to determine not only the amount of circulating DNA in the patient, but also the amount of viral DNA. Papillomavirus detection/genotyping was performed by qPCR. One third of the patients showed at least one high-risk genotype. Conclusion: In this study, differential quantification of both patient and viral circulating DNA was observed.
Keywords: Human papillomavirus; High-risk genotypes; Cervical cancer
Introduction
Cervical cancer is the fourth leading cause of death in the world [1]. This poor prognosis is due to the fact that in two out of three cases the disease is diagnosed in advanced stages. Consequently, advances in detection methods can lead to early diagnosis of the disease and consequently to an improved prognosis. Methods based on tumor markers present in blood are a promising tool for early diagnosis and in general for biochemical screening of patients with cervical cancer [2,3]. They also offer the added value of being non-invasive methods. Evidence has been found of the release of viral DNA into the blood, so its quantification has the potential to serve as a tumor marker. If this viral DNA is detected in the blood, it would indicate possible malignant transformation, which can reflect tumor size, viral load, and response to treatment [4]. Apoptosis is also a natural biological process that leads to cell fragmentation and the release of nuclear material in the form of mono- and oligo-nucleosomes [5]. It has been estimated that several hundred billion to one trillion cells die daily in the adult human body because normal physiology must be replaced by a similar number generated through mitosis.
Despite the existence of an efficient elimination system, a considerable amount of apoptotic genetic material [6] enters the circulation in normal individuals and at elevated levels in a multitude of acute and chronic human pathologies, including cancer [7,8]. Circulating nucleic acid levels increase with age. However, the probable physiological role of circulating nucleic acids in the blood of patients with preneoplastic lesions is beginning to be elucidated [9,10].
Objective
To determine the amount of circulating DNA a viral DNA in the serum of patients with precancerous lesions.
Material and Method
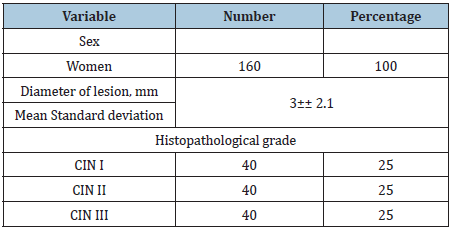
The study was carried out with 40 healthy women and 120 women with precancerous lesions, who attended gynecologicalobstetric care at the Hospital del niño y la Mujer in San Luis Potosí for consultation for gynecological-obstetric care. Of the 120, 40 had cervical intraepithelial neoplasia type I (CINI), 40 had cervical intraepithelial neoplasia type II (CIN II), 40 had cervical intraepithelial neoplasia type III (CINIII). The diagnosis was confirmed by cytology, colposcopy and histopathology. Informed consent was obtained from the patients.
6mL of blood was collected in specific vacutainer tubes for serum separation, containing clot activation. Additive and barrier gel. Separation was performed at room temperature within 2 hours of sample collection. The serum was stored at 4 °C and processed within 6 hours. Blood was separated by centrifugation (1,000×g, 15 min) and passed through a 13mm serum filter (Fisher Scientific) to remove potentially contaminating cells. Twenty ul of proteinase K (20mg/ml, Invitrogen), and the Wizard® Genomic DNA Purification Kit, DNA extraction kit (Promega) were used. DNA was quantified in NanoDrop 2000c UV-Vis for integrity and purity.
On the other hand, papillomavirus detection/genotyping was performed by qPCR in a Pikoreal kit (Thermo Fisher Scientific). Specific oligonucleotides were used to determine the amount of viral DNA, for genotypes 16 and 18, from the same samples (Table 1). The qPCR was performed in 96-well plates, in which were placed: 7 picomoles/microliter of the sense oligo, 5picomoles/ microliter of the antisense oligo, 5 microliters of SYBR Green (Realtime PCR Master Mix), 50 nanograms/microliter of DNA extracted from the samples and adjusted to a10uL final volume with sterile H2O. The plate was placed in the real-time qPCR equipment and programmed to the following conditions (Table 2).
Table 1: Sequences of oligos for E6 HPV-16, E6V PH-18.
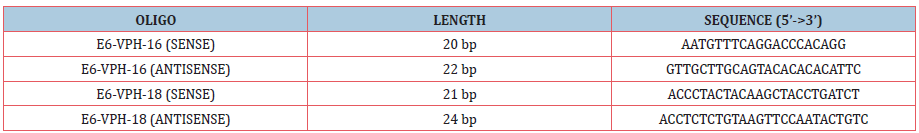
Table 2:Amplification conditions for EVPH-16, EVPH18.

Statistical Analysis
Mean values for healthy women and patients with each stage of precancerous lesion were compared by ANOVA, and with a subsequent Dunnett’s multiple comparison. We also analyzed whether there was a clinicopathological effect using a multiple regression model. The variables indicated for the analyses were entered (p<.05). Multiple linear regression was used to evaluate the possible dependence of mean serum DNA integrity on patient age and clinical information of the lesion. GraphPad Prism version 9.1 was used to perform statistical analysis. A P value of less than 0.05 was considered significant.
Results
Clinical data of healthy subjects and subjects with precancerous lesions
The mean age for healthy subjects was 35 ± 14 years, and that of patients with precancerous lesions was 46 ± 12 years. Table 3 shows the characteristics of the precancerous lesions.
Table 3:
Circulating DNA in serum from patients with precancerous lesions
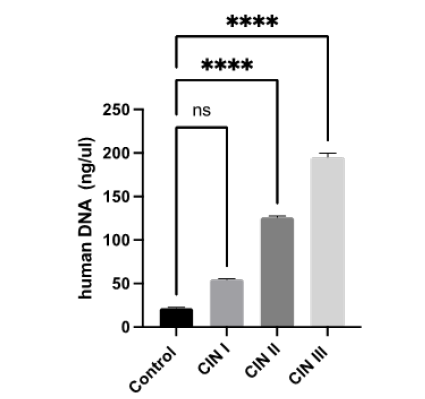
The extracted DNA had a purity between 1.6-1.8 with good integrity. More shDNA (free human DNA) was extracted in samples with CIN III (188ng/ul); CIN II (126ng/ul) and CIN I (55ng/ul), then in serum samples from healthy subjects (controls 20.8ng/ul; Figure 1).
Figure 1:Free human DNA in serum samples taken from controls and patients with pre-cancerous lesions. **** P< 0.0001, with an ANOVA test. CIN, Cervical Intraepithelial Neoplasia; ns, not significative.
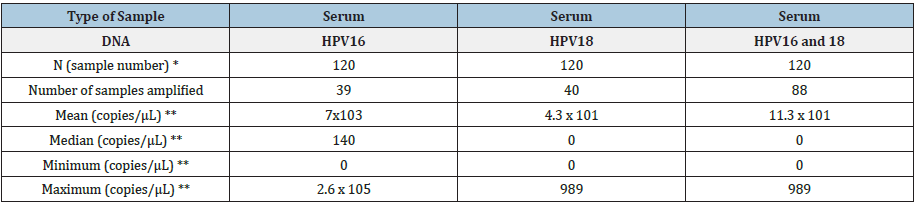
On the other hand, the results obtained for qPCR were as follows: one third (39) of the 120 patients showed at least one high-risk genotype in serum, predominantly HPV 16, with a higher number of copies (700 copies/uL; Figure 2). In addition, more than half of the patients had co-infection with HPV 16 and HPV 18. (Table 4).
Figure 2:Genotyping of HPV 16, and HPV 18, in 10 ul of free DNA from the 120 serum samples collected from patients with pre-cancerous lesions. **, P< 0.001, with an ANOVA test. ns, not significative.
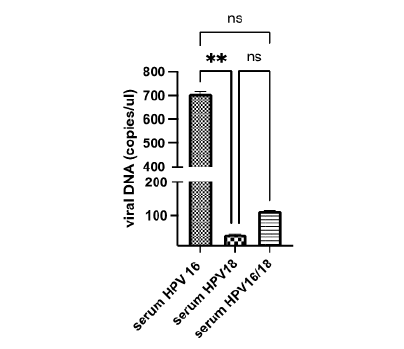
Table 4:
Discussion
Studies have shown elevated levels of free circulating DNA of various forms in serum or plasma from patients with various types of cancers [11-14]. Circulating DNA as a biomarker of malignancies can be detected in the form of allelic imbalance, gene methylation and gene mutation [15]. Recently, it has been shown that the length of cell-free plasma DNA in patients with pancreatic cancer was greater than in healthy controls [16]. Another study has indicated that circulating DNA integrity, calculated from 400bp and 100bp qPCR threshold values of a specific gene, may be a molecular biomarker for the detection of gynecological cancer [17]. To improve our knowledge, this study investigated the significance of free DNA by comparing three groups with precancerous lesions against a control group. All patient samples involved Mexican women from San Luis Potosi capital, and we found an increasing trend as the lesion progresses. It has been documented that the high concentration of free DNA may be due to decreased DNAs an activity, LOH and genetic aberrations, such as allelic imbalance and epigenetic changes of promoter hypermethylation that have been detected in free DNA from patients with other types of cancer such as lung cancer [18]. This may be explained by the fact that DNA released from malignant tumors into the bloodstream has a better diffusion compared to tumor cells [19]. This feature could well correspond to tumor activity or viral load in this case of Human Papilloma virus, which we found to be elevated in patients with CIN III.
Schwarzenbach et al. [20] observed correlations between plasma DNA and circulating tumor cells (CTCs) in prostate cancer patients. They explained this interesting finding by assuming that cell-free tumor DNA in peripheral blood can be derived not only from the primary tumor but also from circulating or disseminated tumor cells. It was found that tumor cells spread throughout the body before the primary tumor has acquired multiple genomic changes and DTCs (disseminated tumor cells) may acquire key molecular changes that may allow them to form overt metastasis. In our case patients with CIN III have genomic damage as we previously reported [3].
Jung et al. [21] reported that 11 of 30 patients with lymph node metastases had increased DNA concentrations associated in only one case with an increased concentration of osteoprotegerin as a distant metastatic indicator in patients with prostate cancer. Therefore, an elevated DNA concentration could indicate a metastatic process already with early lymphatic involvement. This could suggest that CIN III patients with free DNA and HPV viral load are more likely to develop metastasis. The data obtained in the present study are in complete agreement with the majority of investigations that detected higher levels of free DNA in patients with adverse prognosis or increased with metastases for other types of cancer [22,23]. Therefore, serum free DNA, together with molecular typing of HPV type can be considered an important noninvasive and useful biomarker, especially for follow-up in patients with preneoplastic lesions. Our results showed no correlation of any of our markers with clinicopathological parameters. These results are in agreement with previously obtained data [24-26].
However, the quantification of circulating cell-free DNA still has some limitations, such as differences in methodology and lack of standardization in these methodologies, which has hindered the practical implementation of these tumor markers. For example, several studies use plasma to quantify circulating cell- free DNA, while other studies use serum as a template [27].
In addition, some studies performed DNA extraction and measured circulating cell-free DNA levels by qPCR, while other studies use serum or plasma as a direct template to quantify cell-free DNA [28,29]. However, most published reports consider cDNA integrity of serum and/or plasma as a promising molecular biomarker to detect various types of malignancies.
In conclusion, our results revealed a highly significant level of cDNA with high integrity in the plasma of patients with preneoplastic lesion. Therefore, it may become a valuable noninvasive biomarker for the detection and monitoring of preneoplastic lesions, as it provides a simple and inexpensive measure for the detection and follow-up of these lesions. In this study, we observed a differential quantification of circulating DNA in patients, as well as the difference in copy number, with a higher viral load for HPV 16.
References
- IARC (2018) Human Papillomavirus. IARC monographs Eval Carcinog Risks Hum: 1-313.
- Aires GMA, Meireles JRC, Olivera PC, Oliveira JL, Araujo EL, et al. (2011) Micronuclei as biomarkers for evaluating the risk of malignant transformation in the uterine cervix. Genetics and Molecular Research 10(3): 1558-1564.
- Alcántara-Quintana LE, Terán-Figueroa Y, Ortiz-Valdez JA, Gallegos-García V (2018) Micronucleus assay in preneoplastic lesions of cervical cancer. Global Journal for Research Analysis 7(9): 440.
- (2014) American Cancer Society.
- Shen Z (2011) Genomic instability and cancer: An introduction. Journal of Molecular Cell Biology 3(1): 1-3.
- Lowe SW, Lin AW (2000) Apoptosis in cancer. Carcinogenesis 21(3): 485-495.
- Tsai NW, Lin TK, Chen SD, Chang WN, Wang HC, et al. (2011) The value of serial plasma nuclear and mitochondrial DNA levels in patients with acute ischemic stroke. Clin Chim Acta 412(5-6): 476-479.
- Mittra I, Mishra PK, Mansoor S, Samant U, Patkar V, et al. (2010) Abstract LB-103: Circulating chromatin is a novel DNA damaging agent that induces genomic instability and malignant transformation. Cellular and Molecular Biology 70(8).
- Mittra I, Nair NK, Mishra PK (2012) Nucleic acids in circulation: Are they harmful to the host? J Biosci 37(2): 301-312.
- Rekha MR, Pal K, Bala P, Shetty M, Mittra I, et al. (2013) Pullulan-histone antibody nanoconjugates for the removal of chromatin fragments from systemic circulation. Biomaterials 34(27): 6328-6338.
- Leon SA, Shapiro B, Sklaroff DM, Yaros MJ (1977) Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res 37(3): 646-650.
- Anker P, Mulcahy H, Chen XQ, Stroun M (1999) Detection of circulating tumour DNA in the blood (plasma/serum) of cancer patients. Cancer Metastasis Rev 18(1): 65-73.
- Holdenrieder S, Stieber P, von Pawel, Hannelore R, Nagel D, eta l. (2004) Circulating nucleosomes predict the response to chemotherapy in patients with advanced non-small cell lung cancer. Clin Cancer Res 10(18 Pt 1): 5981-5987.
- Herrera LJ, Raja S, Gooding WE, Talal El-Hefnawy, Lori K, et al. (2005) Quantitative analysis of circulating plasma DNA as a tumor marker in thoracic malignancies. Clin Chem 51(1): 113-118.
- Wagner PD, Verma M, Srivastava S (2004) Challenges for biomarkers in cancer detection. Ann NY Acad 1022: 9-16.
- Giacona MB, Ruben GC, Iczkowski KA, Ross TB, Porter DM, et al. (1998) Cell-free DNA in human blood plasma: Length measurements in patients with pancreatic cancer and healthy controls. Pancreas 17(1): 89-97.
- Wang BG, Huang HY, Chen YC, Bristow RE, Keyanunoosh K, et al. (2003) Increased plasma DNA integrity in cancer patients. Cancer Res 63(14): 3966-3968.
- Tissot C, Toffart AC, Villar S, Souquet PJ, Merle P, et al. (2015) Circulating free DNA concentration is an independent prognostic biomarker in lung cancer. Eur Respir J 46(6): 1773- 1780.
- Wu J, Tang W, Huang L, Hou N, Wu J, et al. (2019) The analysis of cell-free DNA concentrations and integrity in serum ofinitial and treated of lymphoma patients. Clin Biochem 63: 59-65.
- Schwarzenbach H, Hoon DS, Pantel K (2011) Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 11(6): 426-437.
- Jung K, Stephan C, Lewandowski M, Klotzek S, Jung M, et al. (2004) Increased cell-free DNA in plasma of patients with metastatic spread in prostate cancer. Cancer Lett 205(2): 173-180.
- Huang A, Zhang X, Zhou SL, Cao Y, Huang XW, et al. (2016) Plasma circulating cell-free DNA integrity as a promising biomarker for diagnosis and surveillance in patients with hepatocellular carcinoma. J Cancer 7(13): 1798-1803.
- Zaher ER, Anwar MM, Kohail HM, El-Zoghby SM, Abo-El-Eneen (2013) Cellfree DNA concentration and integrity as a screening tool for cancer. Indian J Cancer 50(3): 175-183.
- Salvi S, Gurioli G, De Giorgi U, Conteduca V, Tedaldi G, et al. (2016) Cellfree DNA as a diagnostic marker for cancer: Current insights. Onco Targets Ther 9: 6549-6559.
- Catarino R, Ferreira MM, Rodrigues H, Coelho A, Nogal A, et al. (2008) Quantification of free circulating tumor DNA as a diagnostic marker for breast cancer. DNA Cell Biol 27(8): 415-421.
- Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, et al. (2014) Detection of circulating tumor DNA in early- and latestagehuman malignancies. Sci Transl Med 6(224): 224ra24.
- Taback B, Hoon DS (2004) Circulating nucleic acids in plasma and serum: Past, present and future. Curr Opin Mol Ther 6(3): 273-278.
- Taback B, O'Day SJ, Hoon DS (2044) Quantification of circulating DNA in the plasma and serum of cancer patients. Ann N Y Acad Sci 1022: 17-24.
- Jung M, Klotzek S, Lewandowski M (2003) Changes in concentration of DNA in serum and plasma during storage of blood samples. Clin Chem 49(6 Pt 1): 1028-1029.
© 2021. Alcántara-Quintana Luz Eugenia. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






