- Submissions

Full Text
Novel Approaches in Cancer Study
The Role of MicroRNAs in the Progression, Prognostication, and Treatment of Breast Cancer
Chikezie O Madu1, Anming Gu2, Chinua Madu2 and Yi Lu3*
1Departments of Biological Sciences, The University of Memphis, USA, Email: comadu@ memephis.edu
2Department of Biology and Advanced Placement Biology, White Station High School, Memphis, USA, Email: sb108vg@gmail.com
3 Department of Pathology and Laboratory Medicine, University of Tennessee Health Science Center, Memphis, USA, Email: ylu@uthsc.edu
*Corresponding author: Yi Lu, Department of Pathology and Laboratory Medicine,University of Tennessee Health Science Center, Cancer Research Building, Room 258, 19 South Manassas Street, Memphis, TN 38163, USA, Email: ylu@uthsc.edu
Submission: February 25, 2020Published: March 04, 2020

ISSN:2637-773XVolume4 Issue2
Abstract
MicroRNAs (miRNAs) are conserved, small, non-coding RNA molecules, which recently have attracted enormous attention in numerous physiological and pathological conditions. Several studies have shed light on their biogenesis, regulatory mechanisms, and role as effective therapies in diseased conditions. Of interest, miRNA deregulation in numerous cancer types has been researched as potential diagnostic and prognostic tool. Breast cancer (BCa) is the most predominant tumor in women and the main cause of death. Despite advances in diagnosis and new treatments, the death toll from this disease is still higher than many other types of cancer in men and women. A major global health issue plaguing the health and clinical research industry is resistance to BCa treatments. A lot of attention is increasingly directed towards miRNAs as a potential predictor’s response to treatments and as an alternative therapy to BCa treatment. Increasing evidence reveals a fundamental role miRNA plays in cancer development, progression, and treatment. Repeated findings have reinforced evidence of miRNA modulations in breast cancer cells by their effects in cell migration and invasion. Recently, miRNAs have been described as a diagnostic and prognostic tool, which offers promises as biomarkers for advancement of non-invasive and precise methods for screening tumor growth and progression. This review summarizes an overview of miRNA in breast cancer growth and progression, recent applications as biomarkers in a clinical setting in this type of cancers.
Introduction
MicroRNAs (miRNAs)are small single-stranded RNA molecules found in several organisms including plants, animals and some viruses. They are a member of a family of functional RNA molecules referred to as small non-coding RNAs (ncRNAs), which has about 22 nucleotides and functions in RNA silencing and post-transcriptional regulation of gene expression by repressing target gene expression at the translation level [1,2] miRNAs and their roles as regulatory ncRNAs are the most extensively studied member of this class. miRNAs are evolutionarily conserved in both plants and animals and, hence, believed to be fundamental participants in gene regulation right from early life [3-6]. They originate from regions of RNA transcripts that form short hairpins by folding back on themselves. This is one major distinction between these Small interfering RNA (siRNA), which have their origin from longer regions of double-stranded RNA [7].
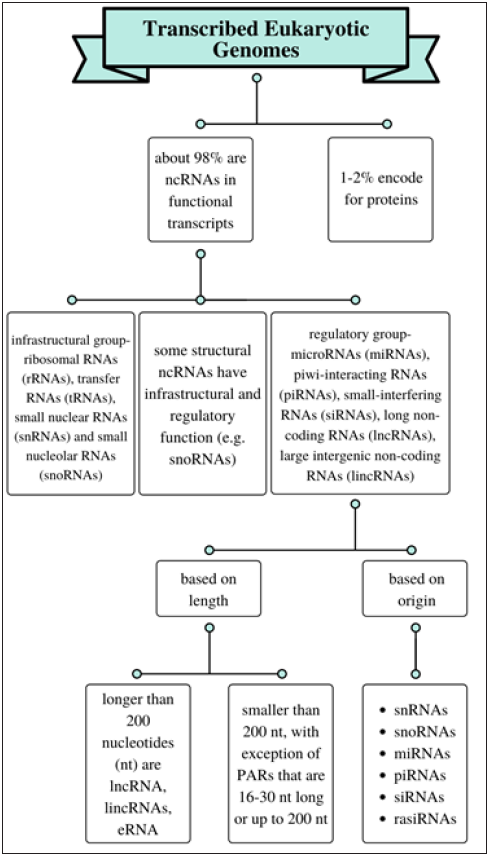
Non-coding RNAs can be categorized into two groups based on their function. Those involved in infrastructure and those that participate in regulatory functions. The regulatory ncRNAs can be further divided in two groups based on their length and origin, and still further as those larger than 200 nucleotides, and those up to 200 or smaller than 200 nucleotides [8-10] (Figure 1). Ambros and his group discovered lin-4 in C. elegans as the first miRNA member in 1993 [1]. and they were observed to repress mRNAs during the translation process that encode proteins of the heterochronic developmental timing pathway in these organism [7] let-7 RNA, a second small RNA was shortly thereafter characterized seven years later as a repressor of lin-41 and also discovered to be conserved in many species. These and several other studies later led researchers to conclude that they might play a related role in regulating the timing of development in humans and other animals [11,12]. Since their description, about 29,000 miRNAs have been discovered different large and microorganisms [13]. Although a little more than 2000 have been discovered in humans, much is still not completely understood regarding their specific cellular function and role in disease progression [14,15]. However, their ability to regulate genes have revealed that they are vital in numerous physiological functions [16] and pathological conditions [17,18], such as early development, cell proliferation, differentiation, apoptosis, disease pathology, tumorigenesis, development and clinical therapy [19,20].
Figure 1: Overview of categorizing RNAs in eukaryotic genome.
miRNAs in humans are largely located in the introns of genes or the ncRNA transcript regions [2] with different groups expressed in different cell types and tissues [21,22]. These small group of gene regulators function by binding to complementary sequences located in the 3’ untranslated region (UTR) of target mRNA, inducing negative regulation [23]. They will induce the degradation of that target when they bind completely to sequences of the complementary mRNAs (an observation commonly noticed in plants). Conversely, a partial binding between a miRNA and its target will lead to translational repression [24]. A specific miRNA molecule can regulate several mRNAs, various miRNAs can work in tandem to regulate a specific mRNA target, and as many as 30% of genes involved in protein coding in humans are mediated by miRNAs [25]. These processes function through an interplay between the miRNA and several signaling molecules, such as cytokines, growth factors, transcription factors, pro-apoptotic and anti-apoptotic genes [26].
The expression of miRNAs is determined by innate cellular factors and different environmental cues [27]. About half of miRNAs can be found in locations prone to tumor formation [28], and abnormally expressed miRNAs have been implicated in most tumors (designated as oncogenic miRNAs-oncomiRs) by acting directly or indirectly on oncogenes and tumor suppressor genes to regulate their expression [19,29,30]. Current research reveals miRNAs as possible tumor biomarkers and are increasingly being studied as novel therapeutic targets. Although most miRNAs are found within the cell, some have also been discovered in extracellular environment such as several biological fluids [31]. This class of miRNAs, generally referred to as circulating miRNAs or extracellular miRNAs, have been identified at high levels in the circulation, and have observed in cell signaling activities with great potential use as non-invasive biomarkers [32-34].
This paper aims to provide a brief overview of the biogenesis and regulation miRNA and describe the mechanisms by which its expression is dysregulated in human diseases in general and specifically in breast cancer- as oncogenes or tumor suppressors. Furthermore, we highlight miRNAs as potential biomarkers in cancer diagnosis, prognosis and treatment and, finally, we discuss some of the research challenges in identifying novel miRNAs and their biological functions and describing their applications in the treatment of diseases.
MicroRNA Biosynthesis
miRNAs synthesis consists of several significant stages. First, RNA polymerase II (and sometimes RNA polymerase III), typically generated from hundreds to several kilobases of pri-miRNA transcripts in the nucleus [35-37] structurally similar to matured transcripts but with a structural stem loop [36,38]. The pri-miRNAs then forms a microprocessor complex by interacting with the ribonuclease Drosha and double stranded RNA binding protein, DiGeorge syndrome critical region gene 8 (DGCR8), which cleaves it to generate a ~ 85-nucleotidestem–loop structure called precursor miRNA (pre-miRNA) hairpin [39,40].
Subsequent to this, the pre-miRNA is exported to the cytoplasm by Exportin 5 (the nucleocytoplasmic shuttle) where the ribonuclease Dicer complex and transactivation-responsive RNA-binding protein 2 (TARBP2) cleaves the loop, converting the pre-miRNA into a double-stranded miRNA duplex [41]. Following a strand separation, one of the strands of the miRNA duplex binds onto Argonaute (a protein family that binds different classes of small non-coding RNAs) to create the miRNA-Induced Silencing Complex (miRISC), while the other strand gets degraded [42-46] (Figure 2).
Figure 1: The biosynthesis pathway for miRNAs.
Note: RNA polymerase II is usually involved in transcribing miRNA genes, producing large primary transcripts
called pri-miRNAs. The pri-miRNAs are cleaved by a microprocessor complex into a stem–loop structure called
pre-miRNA. The pre-miRNAs are processed by another RNase III enzyme Dicer to a ~ 20–22-nucleotide miRNA/
miRNA* duplex after they are transported from the nucleus to the cytoplasm by Ran/GTP/Exportin 5 complex.
Following the separation of the duplex, the mature miRNA is combined with a protein complex termed RISC,
which can then mediate gene silencing by cleavage and degradation, or repressing translation [47].

This newly formed complex is now set for down-regulating gene expression by either perfectly binding all along their length to target complementary mRNA transcripts-degrading the transcript, or more frequently, partially binding with complementary transcripts- inhibiting translation [1,47] (Figure 2).Although the primary mechanism of miRNA action in mammals is thought to be as described above, it has several functional roles [9]. Usually, in animals, it’s key mechanism of action involves reducing mRNA translation, and individual miRNA can subsequently inhibit the translation of over 200 target genes [7,48-50]. Additionally, it is purported that some miRNAs have other activities other than post-transcriptional suppression, for instance some studies have revealed that some may epigenetically regulate transcription by targeting promoters. Astonishingly, they have also been observed under specific circumstances to increase translation and, consequently, up-regulate gene expression [9], miRNAs can also frequently initiate histone modification and DNA methylation of promoter regions, thereby affecting the expression of specific genes [51,52]. These diverse actions can create a state that miRNAs confer upon them ability to play integral roles in normal cellular functions [53-58] and, if dysregulated, disease [59].
The Role of MiRNAs in Cancers and Other Human Diseases
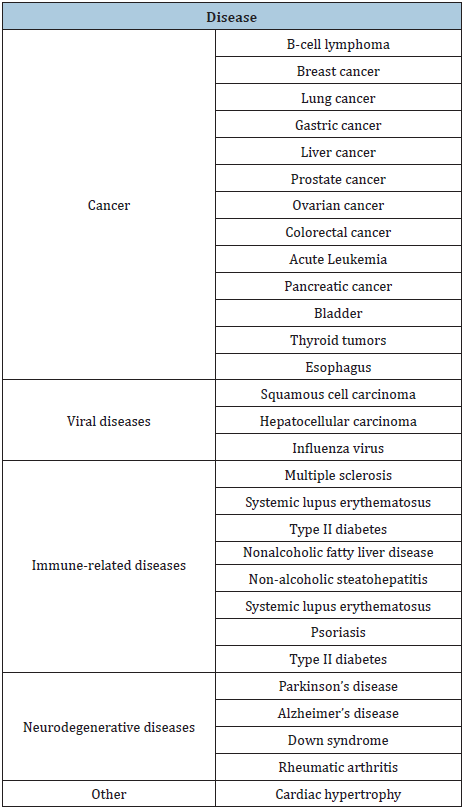
Calin et al. [60] were the first to reveal a link between miRNA s and human disease when they reported in 2002 that deregulation of certain miRNA (miR-15 and miR-16) results in B-cell chronic lymphocytic leukemia (CLL). Eventually, many other miRNAs came to be linked to cancer incidence [61] because of altered expression of miRNA through such actions like DNA amplification, deletion and mutations on specific miRNA loci, epigenetic silencing or inhibition of certain miRNA processing [62]. Subsequently, it has been reported that other diseases could arise from the mechanisms just listed, dysfunction of miRNA biogenesis and dysregulation of miRNAs and their targets [63]. Currently, several other instances where miRNA genes are either deleted or amplified in tumors have been compiled from numerous studies [64] and concluded that miRNAs can be implicated in numerous human diseases, including myocardial infarction and cardiovascular diseases [65-68] and cancer [58] (Table 1).
Although almost 100 miRNA-linked diseases have been described [63,64], scientists are still trying to better understand the mechanisms of miRNAs in diseases and the patterns of involvement. Lu M, et al. [63] in their analysis of associations between human microRNA and disease, observed a negative correlation between microRNA tissue specificity and the number of diseases associated with it. They also reported a link between microRNA conservation and disease. This conservation is also linked with the susceptibility to a human disease, further unraveling the roles of miRNA in diseases.
Further, the researchers stated that the fundamental mechanisms underlying respective diseases might be different, although this different group of diseases may be linked to the same miRNA. A phenomenon they suspect might be due to a correlation between miRNA tissue specificity and a disease [63]. A dysfunction in members of the miRNA family would show a similar phenotype because they may possess analogous functions and participate in the same biological processes [69,70]. Finally, usual factors, under identical circumstance and function, can regulate miRNAs in surrounding cells, and an abnormality in these functions could cause same disease [63].
There is adequate evidence to show that miRNAs are key players in the initiation and progression of human cancer [71]. The deregulation of miRNA in various cancer sub types has been widely researched since Calin et al. [60] first showed the biological role of miRNAs in cancer development. miRNAs have been identified to be linked to numerous and different cancers including lung, breast, and prostate, ovarian, colorectal, acute leukemia, pancreatic, squamous cell carcinoma hepatocellular carcinoma, and several others [69,72-74] Table 1.
Table 1: Somatic diseases and concomitant OMD.
These discoveries, subsequently, has led to increase attention and studies in the downregulation of miRNAs in cancer tissues compared with normal tissues. Also, there are some evidence that miRNAs in stress conditions, such as hypoxia, stimulate some targets [75-76]. For instance, miR-210 may be involved as a vital player in HIF-1a-induced macrophage necroptosis [77]. Intriguingly, several of these studies have revealed that all cancers display unique microRNAs [78,79]. The abnormal expression of miRNA in tumors has led scientists to conclude that the dysregulated miRNAs could influence one or numerous hallmarks of cancer initiation and progression. MiRNAs dysregulation in human cancers (which can be at the transcriptional or translational level) acts as either oncogenes or tumor suppressors, depending upon their target transcripts. Table 2 shows the hallmarks of breast cancer associated with the deregulation of several respective miRNAs [80]. For instance, miR-155, has been shown to be upregulated in several hematopoietic malignancies and tumors of the breast, lung and pancreas [81,82] and miR-29 functions as a tumor suppressor through its pro-myogenic function [83].
Table 2: miRNAs as key regulators of Breast cancer hallmarks. Expression of miRNAs (up-regulated and down-regulated) grouped according to their function in the hallmarks of breast cancer.
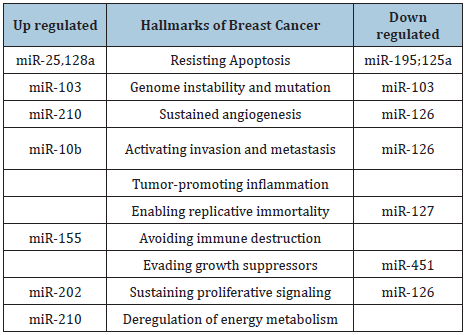
The major distinction between the mechanism of action of oncogenic miRNAs (OncomiRs) and tumor suppressor miRNAs is that while the former represses the expression of tumor suppressor genes and are often upregulated in cancer, the latter targets oncogenes and are often downregulated in cancer [84-88]. For example, one of the most overexpressed miRNAs in human epithelial malignancies, miRNA-21 (miR-21), downregulates numerous tumor suppressors [89], and some studies have suggested the involvement of the tumor suppressor gene, p53, in the regulation of miRNAs [90], and miR-506 has been shown to induce apoptosis in cervical cancer cells, through its direct target hedgehog pathway transcription factor. [91] Although, many studies have revealed several miRNAs that function in both ways- oncogenic or tumor suppressive abilities, depending on tumor type and cellular context [92]. Furthermore, miRNAs have been observed to be involved in tumor metastasis [93] and because miRNAs exhibit impressive tissue specificity (and patterns of gene activity), it is often possible to establish the tissue of origin of a metastasis in patients with unidentified primary tumors [78,94].
Role of miRNAs in Breast Cancer Progression and Treatment
Breast cancer is the second most commonly diagnosed cancer among American women after skin cancer, and fifth most common cause of cancer mortality worldwide. In the United States, there were about 266,120 new cases of invasive breast diagnosed in the women and 2,550 cases diagnosed in men 2018. In that year, an estimated 41,400 breast cancer deaths were estimated, 480 men and more than 3.5 million survivors [95]. The treatment of this disease has proven to be very challenging due to its multifaceted and heterogeneous nature, comprising of numerous molecular subtypes, each with diverse respective cellular origin, treatment response, and clinical outcome [96,97].
Various genetic and epigenetic changes in these cells activate the various features of cancer (sustaining proliferative signaling, evading growth suppressors, resistance to cell death, immortality, angiogenesis, stimulating invasion and metastasis, reprogramming energy metabolism, and escaping immune destruction), which all leads to malignant breast tumor progression [98]. Recent studies have implicated noncoding RNAs (ncRNAs), like MiRNAs, as sources of epigenetic alteration, genetic variations, modifications in miRNA biogenesis, and transcriptional repression [8,99]. An example of an oncogenic miRNA associated with breast cancer is MiR-21, which is one of the most highly expressed miRNAs in breast cancer. It has been shown to inhibit several tumor-suppressor genes and consequently promotes cell growth and invasion, and tumor metastasis, and its upregulation is associated with poor prognosis [89]. MiR-155, another oncogenic miRNA, is known to target, a human breast cancer susceptibility gene (BRCA1) involved in DNA repair and cell cycle progression and thereby regulate several signaling pathways linked to the cell’s growth and survival [100-102] miR-335, an example of a tumor-suppressor, influences vital cellular functions such as proliferation and apoptosis in breast cancer cells [103] miRNAs are involved also in breast cancer metastasis, one of the key factors associated with mortality [95].
To become metastatic, tumor cells must acquire epithelialmesenchymal transition (EMT) and angiogenesis [104] and several miRNAs target mechanisms that promote these processes in breast tumors [105], for instance by inducing phenotypic changes related to the development of metastasis [106]. One of such is miR-205, which target genes’ expression vascular endothelial growth factor’s A(VEGF-A) that has been linked to breast cancer progression migration and metastasis), have proven to promote breast carcinoma survival [107]. In addition to promoting metastasis, certain miRNAs have also been linked to different factors that affect breast cancer prognosis. For instance, studies have reported that the expression levels of miR-210 were correlated with tumor aggressiveness and poor prognosis, and therefore can be associated with breast cancer development [108,109]. Other examples include miR-9 [110,111] which may be useful as biomarkers for predicting metastasis and the possibility of the tumor recurring- a negative prognosis of patients with breast cancer.
miRNA expression is deregulated in cancer and the physical appearance of these cells are often altered by targeting the expression. These discoveries have thereby established the basis for the use of miRNAs in anticancer therapy [112]. The two-prong approach employed in using miRNA in anticancer therapy is either by inhibiting the function of miRNAs as oncogenes or upregulating the expression of their tumor-suppressive functions [113]. One main approach employed in blocking the expression is by a direct strategy, which involves either blocking the expression of oncogenic miRNAs or substituting for the loss of expression of a tumor suppressor miRNA through the use of oligonucleotides or virusbased constructs. Another strategy, an indirect approach, targets their transcription and processing by altering the expression of the miRNA with the use of drugs [114].
Certain miRNAs have been used to successfully perform a wide- ranging silencing of cancer pathways due to their ability to target those genes from identical pathways and those from cross linking pathways. miR-205 was one of the miRNAs discovered to be a potential compound in breast cancer therapeutics [115]. Early studies revealed that miR-181a possesses an anti-breast cancer function by inhibiting tumor invasion and metastasis, has the ability to induce cancer cell death, and improve drug sensitivity. However, it was observed in studies that downregulation is more aggressive or late stage breast cancer, thereby functioning as a tumor suppressor gene [116-118]. A benefit of using miRNAs as the anti-cancer therapy includes the fact that they are produced in human cells unlike other synthetic compounds like chemotherapies. Another advantage is that microRNAs target multiple genes from the same pathway [119].
The first miRNA drug developed was by the company Santaris Pharma A/S. It was an LNA-modified oligonucleotide, SPC3649 used to repress the expression of miR-122, in treating chronic hepatitic C virus (HCV) infection [120]. An understanding of miRNAs as oncogenes and/or tumor suppressors in cancers, has paved the way for researchers to study their potential use as therapeutic targets for clinical application and as biomarkers for cancer detection and development. miRNAs have been considered a vital candidate for biomarkers because they have some advantages over candidates such as proteins and metabolites, for instance, they are stable enough to resist several enzymes in body [121].
MicroRNAs as Potential Biomarkers in Cancer Diagnostic
Detecting and diagnosing breast cancer early in patients offers more favorable outcomes in treating the disease [122] and clinicians and researchers are increasingly relying on the use of biomarkers to detect early stages of breast cancer. Biomarkers have prognostic value; used in predicting patient’s outcome, disease advancement and relapse, despite ongoing treatment. It is therefore not surprising that several studies, both past and ongoing, involve a search for viable prognostic biomarkers. miRNAs have been linked with clinical and pathological features, making them ideal for prognostic purposes in several tumor categories and subcategories of breast cancer patients [106,123,].
The discovery that miRNAs can target various genes and modify numerous pathways and hence target gene expression, made them attractive candidates as diagnostic, prognostic, or predictive breast cancer biomarkers in recent years [124]. For instance, several studies have reported that the expression of miR-181a is correlated with of clinic pathological features of breast tumors [105,125-128] and in prostate cancer patients, miR-141 is upregulated [129].
Additionally, miRNA biomarkers have a greater potential to reveal early diagnosis and are easier to analyze using biotechnology methods because of their locations upstream in regulation cascades. miRNAs are released into circulation from the primary tumor and other cell types. They are then secreted into the extracellular space, from there they move into the circulating body fluid, and can eventually be detected in almost all fluids in the body including the whole blood, serum, plasma, saliva, urine, and several others. [130]. Although biomarkers can be measured in tissues, body fluids, or both, researchers prefer, when evaluating healthy individuals, to analyze body fluid. Thus, circulating serum miRNAs have been speculated as a potential, non-invasive diagnostic and prognostic biomarkers for cancer and other diseases such as, diabetes mellitus and neurological disorders [131-133].
However, the total amount of distinct miRNAs is considerably different among the numerous fluids, with the highest detected in saliva, breast milk, and seminal fluid the highest; the lowest in human urine [129,130,134-136]. Non-invasive miRNA biomarkers are more attractive because they present fewer complications linked to the collection of specimens. Due to certain intrinsic characteristic they possess, these circulating miRNAs are ideal candidates for non-invasive biomarkers. circulating miRNAs have been detected in various disease conditions, and, when detected in the peripheral blood circulation and other body fluids could indicate disease progression [137-139]. Some circulating miRNAs can also function as prognostic biomarkers in breast cancer patients, and numerous research studies have reported their possible use in diagnosing metastasis [140].
The use of circulating miRNAs as breast Cancer biomarkers has been plagued with some challenges. One major challenge is their low abundance, which made their detection difficult using microarrays or other basic miRNA profiling techniques [141]. However, scientists have come up with several methods to overcome these obstacles. Another main challenge in the analysis of miRNAs in body fluids is the need for an internal normalization. Certain genes, often used as internal normalization during miRNA analysis in cells and tissues, are not feasible in serum analysis since these controls are easily degraded and not detected in serum [142]. Further research is still required to ascertain the specificity of the microRNA biomarkers for the respective type of cancer in addition to methods to standardize collection and analyze samples. Also, a broad profiling data of circulating miRNA during chemotherapy treatment will be very essential to identify novel circulating markers of response to treatment.
Conclusion
The survival and clinical outcome of breast cancer patients following treatment has been linked to the heterogeneity of the disease. Numerous past research studies in this field has focused on understanding the mechanism involved in initiating and developmental of phenotypic alterations that occur on the epithelial tissue of the breast gland, and although many culprits have been identified as active participants in this process, recent studies coming out have revealed the strong involvement that miRNAs plays in the regulation of breast cancer- either as suppressors or initiators of tumor development. Also, miRNAs have been shown to be potential reliable prognostic biomarkers for breast cancer progression and overall survival, which could be useful in monitoring patient’s progress and the identification of effective treatments.
Research has taken a long stride in the area of improving breast cancer prognostication and numerous mRNA-based tests are presently available towards that goal; however, its clinical use still presents some limitations. This calls for new biomarkers that will provide more accurate monitoring and revelation of treatment protocols and therapeutics, as well as follow-up plans on a personalized basis. Current research in this area seem to be mainly preclinical and, therefore, inconclusive from patient cohorts. Although the results thus far are encouraging, a lot of study still persists in identifying miRNAs as true predictors of success or failure in individual patient’s treatment.
Despite the huge potential of miRNA research in vitro, and its promise to advance precision medicine in breast cancer, more elaborate in vivo models will greatly help to better establish the roles of individual miRNAs and their families. Furthermore, a broad comprehension of their mechanism of action and the pathways they regulate can be acquired through enhanced experimental approaches that will identify specific and critical miRNA targets involved in cancer and determine their respective contribution to tumor transformation.
Although hundreds of different microRNAs have been currently described that can identify and act on specific genes in multiple ways, extensive studies are still needed to safely conclude and state the general function of microRNAs in breast cancer development. Therefore, it may be too premature to predict the total implication of the role of miRNAs in cancer, in general, and breast cancer in particular.
Conflicts of Interest
None to declare.
Acknowledgment
We thank Ava Wang for generating the figures used in this paper.
References
- Lee RC, Feinbaum RL, Ambros V (1993) The elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75(5): 843-854.
- Ambros V (2004) The functions of animal microRNAs. Nature 431(7006): 350-355.
- Axtell MJ, Bartel, DP (2005) Antiquity of microRNAs and their targets in land plants. The Plant Cell 17(6): 1658-1673.
- Tanzer A, Stadler PF (2004) Molecular evolution of a microRNA cluster. J Mol Biol 339(2): 327-335.
- Chen K, Rajewsky N (2007) The evolution of gene regulation by transcription factors and microRNAs. Nature Reviews Genetics 8(2): 93-103.
- Lee CT, Risom T, Strauss WM (2007) Evolutionary conservation of microRNA regulatory circuits: An examination of microRNA gene complexity and conserved microRNA-target interactions through metazoan phylogeny. DNA Cell Biol 26(4): 209-218.
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2): 281-297.
- Esteller M (2011) Non-coding RNAs in human disease. Nat Rev Genet 12(12): 861-874.
- Kaikkonen MU, Lam MT, Glass CK (2011) Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc Res 90(3): 430-440.
- Boštjančič E, Glavac D (2013) MicroRNAs and lncRNAs as tumour suppressors, future aspects of tumor suppressor gene. InTech.
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, et al. (2002) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403(6772): 901-916.
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, et al. (2000) Conservation of the sequence and temporal expression of let 7 heterochronic regulatory RNA. Nature 408(6808): 86-89.
- Cho WC (2007) OncomiRs: The discovery and progress of microRNAs in cancers. Mol Cancer 6: 60.
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2006) miRBase: Tools for microRNA genomics. Nucleic Acids Res 36: D154-158.
- Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, et al. (2005) Phylogenetic shadowing and computational identification of human microRNA genes. Cell 120(1): 21-24.
- Hwang HW, Mendell JT (2006) MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer 94(6): 776-780.
- Junn E, Mouradian MM (2012) MicroRNAs in neurodegenerative diseases and their therapeutic potential. Pharmacol Ther 133(2): 142-150.
- Maccani MA, Padbury JF, Marsit CJ (2011) miR-16 and miR-21 expression in the placenta is associated with fetal growth. PLoS ONE 6(6): e21210.
- Wei R, Yang J, Liu GQ, Gao MJ, Hou WF, et al. (2013) Dynamic expression of microRNAs during the differentiation of human embryonic stem cells into insulin-producing cells. Gene 518(2): 246-255.
- Ono K, Horie T, Nishino T, Baba O, Kuwabara Y, et al. (2015) MicroRNA-33a/b in lipid metabolism- novel "thrifty" models. Circ J 79(2): 278-284.
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, et al. (2002) Identification of tissue-specific microRNAs from mouse. Curr Biol 12(9): 735-739.
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, et al. (2005) MicroRNA expression in zebrafish embryonic development. Science 309(5732): 310-311.
- Bartel DP (2009) MicroRNAs: Target recognition and regulatory functions. Cell 136(2): 215-233.
- Dykxhoorn DM (2010) MicroRNAs and metastasis: little RNAs go a long way. Cancer Res 70(16): 6401-6406.
- Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120(1): 15-20.
- Chen CZ (2005) MicroRNAs as oncogenes and tumor suppressors. N Engl J Med 353(17): 1768-1771.
- Paranjape T, Slack FJ, Weidhaas JB (2009) MicroRNAs: Tools for cancer diagnostics. Gut 58(11): 1546-1554.
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, et al. (2004) Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA 101(9): 2999-3030.
- Ma L, Teruya-Feldstein J, Weinberg RA (2007) Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449(7163): 682-688.
- Farazi TA, Hoell JI, Morozov P, Tuschl T (2013) MicroRNAs in human cancer. Adv Exp Med Biol 774: 1-20.
- Sohel MH (2016) Extracellular/circulating microRNAs: release mechanisms, functions and challenges. Achiev Life Sci 10(2): 175-186.
- Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, et al. (2011) MicroRNAs in body fluids-the mix of hormones and biomarkers. Nat Rev Clin Oncol 8(8): 467-477.
- Schwarzenbach H, Nishida N, Calin GA, Pantel K (2014) Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol 11(3): 145-156.
- Aleckovic M, Kang Y (2015) Regulation of cancer metastasis by cell-free miRNAs. Biochim Biophys Acta 1855(1): 24-42.
- Lee Y, Kim M, Han J, Yeom KH, Lee S, et al. (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23(20): 4051-4060.
- Zhou X, Ruan J, Wang G, Zhang W (2007) Characterization and identification of microRNA core promoters in four model species. PLoS Comput Biol 3(3): e37.
- Borchert GM, Lanier W, Davidson BL (2006) RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol 13(12): 1097–1101.
- Cai X, Hagedorn CH, Cullen BR (2004) Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10(12): 1957-1966.
- Lee Y, Ahn C, Han J, Choi H, Kim J, et al. (2003) Provost P, Rådmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature 425(6956): 415-419.
- Gregory RI, Chendrimada TP, Shiekhattar R (2006) MicroRNA biogenesis: isolation and characterization of the microprocessor complex. Methods Mol Biol 342: 33-47.
- Lund E, Dahlberg JE (2006) Substrate selectivity of exportin 5 and dicer in the biogenesis of microRNAs. Cold Spring Harb Symp Quant Biol 71: 59-66.
- Murchison EP, Hannon GJ (2004) miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr Opin Cell Biol 16(3): 223-239.
- Schwarz DS, Zamore PD (2002) Why do miRNAs live in the miRNP? Genes Dev 16(9): 1025-1031.
- Krol J, Sobczak K, Wilczynska U, Drath M, Jasinska A, et al. (2004) Structural features of microRNA (miRNA) precursors and their relevance to miRNA biogenesis and small interfering RNA/short hairpin RNA design. J Biol Chem 279(40): 42230-42239.
- Khvorova A, Reynolds A, Jayasena SD (2003) Functional siRNAs and miRNAs exhibit strand bias. Cell 115(2): 209-216.
- Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, et al. (2003) Asymmetry in the assembly of the RNAi enzyme complex. Cell 115(2): 199-208.
- Lee RC, Feinbaum RL, Ambros V (1993) The elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75(5): 843-854.
- Pillai RS (2005) MicroRNA function: Multiple mechanisms for a tiny RNA? RNA 11(12): 1753-1761.
- Pillai RS, Bhattacharyya SN, Filipowicz W (2007) Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol 17(3): 118-126.
- Ying SY, Chang DC, Lin SL (2008) The microRNA (miRNA): overview of the RNA genes that modulate gene function. Mol Biotechnol 38(3): 257-268.
- Tan Y, Zhang B, Wu T, Skogerbø G, Zhu X, et al. (2009) Transcriptional inhibiton of Hoxd4 expression by miRNA-10a in human breast cancer cells. BMC Mol Biol 10(1): 12.
- Hawkins PG, Morris KV (2008) RNA and transcriptional modulation of gene expression. Cell Cycle 7(5): 602-607.
- Bartel DP, Chen CZ (2004) Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet 5(5): 396-400.
- Hobert O (2007) miRNAs play a tune. Cell 131(1): 22-44.
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, et al. (2007) A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129(7): 1401-1414.
- Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW (2009) A microRNA imparts robustness against environmental fluctuation during development. Cell 137(2): 273-282.
- Mendell JT (2008) miRiad roles for the miR-17-92 cluster in development and disease. Cell 133(2): 217-222.
- Croce CM (2009) Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 10(10): 704-714.
- Mraz M, Pospisilova S (2012) MicroRNAs in chronic lymphocytic leukemia: From causality to associations and back. Expert Rev Hematol 5(6): 579-581.
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, et al. (2002) Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 99(24): 15524-15529.
- Musilova K, Mraz M (2014) MicroRNAs in B cell lymphomas: How a complex biology gets more complex. Leukemia 29(5): 1004-1017.
- Alajez NM, Lenarduzzi M, Ito E, Hui AB, Shi W, et al. (2011) MiR-218 suppresses nasopharyngeal cancer progression through downregulation of survivin and the SLIT2-ROBO1 pathway. Cancer Res 71(16): 2381-2391.
- Lu M, Zhang Q, Deng M, Miao J, Guo Y, et al. (2008) An analysis of guman microRNA and disease associations. PLoS One 3(10): e3420.
- Jiang Q, Wang Y, Hao Y, Juan L, Teng M, et al. (2009) miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res 37: D98-D104.
- Eulalio A, Mano M, Dal Ferro M, Zentilin L, Sinagra G, et al. (2012) Functional screening identifies miRNAs inducing cardiac regeneration. Nature 492(7429): 376-381.
- Quiat D, Olson EN (2013) MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment. J Clin Invest 123(1): 11-18.
- Hilton C, Neville MJ, Karpe F (2012) MicroRNAs in adipose tissue: their role in adipogenesis and obesity. Int J Obes 37(3): 325-332.
- Dehwah MA, Xu A, Huang Q (2012) MicroRNAs and type 2 diabetes/obesity. J Genet Genomics 39(1): 11-18.
- Li Y, Kowdley KV (2012) MicroRNAs in common human diseases. Genomics Proteomics Bioinf 10(5): 246-253.
- Ardekani AM, Naeini MM (2010) The role of microRNAs in human diseases. Avicenna J Med Biotechnol 2(4): 161-179.
- Taylor MA, Schiemann WP (2014) Therapeutic opportunities for targeting microRNAs in cancer. Mol Cell Ther 2(30): 1-13.
- Rothschild SI, Tschan MP, Federzoni EA, Jaggi R, Fey MF, et al. (2012) MicroRNA-29b is involved in the Src-ID1 signaling pathway and is dysregulated in human lung adenocarcinoma. Oncogene 31(38): 4221-4232.
- Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, et al. (2008) Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451(7175): 147-152.
- Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, et al. (2007) MicroRNA expression profiling in prostate cancer. Cancer Res 67(13): 6130-6135.
- Vasudevan S, Tong Y, Steitz JA (2007) Switching from repression to activation: microRNAs can up-regulate translation. Science 318(5858): 1931-1934.
- Leung AK, Sharp PA (2007) MicroRNAs: A safeguard against turmoil? Cell 130(4): 581-585.
- Karshovska E, Schober A (2017) HIF-1a promotes necroptosis in macrophages by micrornas. Atherosclerosis 263: e11.
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, et al. (2005) MicroRNA expression profiles classify human cancers. Nature 435(7043): 834-838.
- Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6(11): 857-866.
- Goh JN, Loo SY, Datta A, Siveen KS, Yap WN, et al. (2016) microRNAs in breast cancer: regulatory roles governing the hallmarks of cancer. Biol Rev Camb Philos Soc 91(2): 409-428.
- Eis PS, Tam W, Sun L, Chadburn A, Li Z, et al. (2005) Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA 102(10): 3627-3632.
- Kluiver J, Poppema S, de Jong D, Blokzijl T, Harms G, et al. (2005) BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol 207(2): 243-249.
- Wang H, Garzon R, Sun H, Ladner KJ, Singh R, et al. (2008) NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell 14(5): 369-381.
- Zhang JG, Wang JJ, Zhao F, Liu Q, Jiang K, et al. (2010) MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC). Clin Chim Acta 411(11-12): 846-852.
- Liu M, Tang Q, Qiu M, Lang N, Li M, et al. (2011) miR-21 targets the tumor suppressor RhoB and regulates proliferation, invasion and apoptosis in colorectal cancer cells. FEBS Lett 585(19): 2998-3005.
- Zhu S, Wu H, Wu F, Nie D, Sheng S, et al. (2008) MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res 18(3): 350-359.
- Li X, Xin S, He Z, Che X, Wang J, et al. (2014) MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor PDCD4 and promotes cell transformation, proliferation, and metastasis in renal cell carcinoma. Cell Physiol Biochem 33(6): 1631-1642.
- Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, et al. (2008) Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem 283(2): 1026-1033.
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, et al. (2005) A microRNA polycistron as a potential human oncogene. Nature 435(7043): 828-833.
- Wen SY, Lin Y, Yu YQ, Cao SJ, Zhang R, et al. (2014) miR-506 acts as a tumor suppressor by directly targeting the hedgehog pathway transcription factor Gli3 in human cervical cancer. Oncogene 34(6): 717-725.
- Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, et al. (2007) Altered microRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res 67(24): 11612-11620.
- Ma L, Weinberg RA (2008) Micromanagers of malignancy: role of microRNAs in regulating metastasis. Trends Genet 24(9): 448-456.
- Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, et al. (2008) MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol 26(4): 462-469.
- ACS: Atlanta. Breast cancer facts & figures 2017-2018.
- Jema A, Siegel R, Xu J, Ward E (2010) Cancer statistics. Cancer J Clin 60(5): 277-300.
- Cardoso F, Harbeck N, Barrios CH, Bergh J, Cortés J, et al. (2016) Research needs in breast cancer. Ann Oncol 28(2): 208-217.
- Byler S, Goldgar S, Heerboth S, Leary M, Housman G, et al. (2014) Genetic and epigenetic aspects of breast cancer progression and therapy. Anticancer Res 34(3): 1071-1077.
- Bertoli G, Cava C, Castiglioni I (2015) MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics 5(10): 1122-1143.
- Peng Y, Croce CM (2016) The role of MicroRNAs in human cancer. Signal Transduct Target Ther 1: 15004.
- Faraoni I, Antonetti FR, Cardone J, Bonmassar E (2009) miR-155 gene: a typical multifunctional microRNA. Biochim Biophys Acta 1792(6): 497-505.
- O'Donovan PJ, Livingston DM (2010) BRCA1 and BRCA2: breast/ovarian cancer susceptibility gene products and participants in DNA double-strand break repair. Carcinogenesis 31(6): 961-967.
- Szabo CI, King MC (1995) Inherited breast and ovarian cancer. Hum Mol Genet 4 Spec No: 1811-1817.
- Heyn H, Engelmann M, Schreek S, Ahrens P, Lehmann U, et al. (2011) MicroRNA miR-335 is crucial for the BRCA1 regulatory cascade in breast cancer development. Int J Cancer 129(12): 2797-2806.
- Geiger TR, Peeper DS (2009) Metastasis mechanisms. Biochim Biophys Acta 1796(2): 293-308.
- Taylor MA, Sossey-Alaoui K, Thompson CL, Danielpour D, Schiemann WP (2013) TGF-beta upregulates miR-181a expression to promote breast cancer metastasis. J Clin Invest 123(1): 150-163.
- Madhavan D, Zucknick M, Wallwiener M, Cuk K, Modugno C, et al. (2012) Circulating miRNAs as surrogate markers for circulating tumor cells and prognostic markers in metastatic breast cancer. Clin Cancer Res 18(21): 5972-5982.
- Hu Y, Qiu Y, Yagüe E, Ji W, Liu J, et al. (2016) miRNA-205 targets VEGFA and FGF2 and regulates resistance to chemotherapeutics in breast cancer. Cell Death Dis 7(6): e2291.
- Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, et al. (2008) hsa-miR-210 is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res 14(5): 1340-1348.
- Rothe F, Ignatiadis M, Chaboteaux C, Haibe-Kains B, Kheddoumi N, et al. (2011) Global microRNA expression profiling identifies MiR-210 associated with tumor proliferation, invasion and poor clinical outcome in breast cancer. PLoS ONE 6(6): e20980.
- Ma L, Young J, Prabhala H, Pan E, Mestdagh P, et al. (2010) miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol 12(3): 247-256.
- Yu Z, Willmarth NE, Zhou J, Katiyar S, Wang M, et al. (2010) microRNA 17/20 inhibits cellular invasion and tumor metastasis in breast cancer by heterotypic signaling. Proc Natl Acad Sci USA 107(18): 8231-8236.
- Garzon R, Calin GA, Croce CM (2009) MicroRNAs in cancer. Annual Rev Medicine 60:167-179.
- Ha TY (2011) MicroRNAs in human diseases: from cancer to cardiovascular disease. Immune Netw 11(3): 135-154.
- Ling H, Fabbri M, Calin GA (2013) MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov 12(11): 847-865.
- Smith L, Baxter EW, Chambers PA, Green CA, Hanby AM, et al. (2015) Down-regulation of miR-92 in breast epithelial cells and in normal but not tumor fibroblasts contibutes to breast carcinogenesis. PLoS One 10(10): e0139698.
- Berber U, Yilmaz I, Narli G, Haholu A, Kucukodaci Z, et al. (2014) MiR-205 and miR-200c: predictive micro RNAs for lymph node metastasis in triple negative breast cancer. J Breast Cancer 17(2): 143-148.
- Wang B, Li J, Sun M, Sun L, Zhang X (2014) MiRNA expression in breast cancer varies with lymph node metastasis and other clinicopathologic features. IUBMB Life 66(5): 371-377.
- Jiao X, Zhao L, Ma M, Bai X, He M, et al. (2013) MiR-181a enhances drug sensitivity in mitoxantone-resistant breast cancer cells by targeting breast cancer resistance protein (BCRP/ABCG2). Breast Cancer Res Treat 139(3): 717-730.
- Giza DE, Vasilescu C, Calin GA (2014) Key principles of miRNA involvement in human diseases. Discoveries (Craiova) 2(4): e34.
- Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, et al. (2010) Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327(5962): 198-201.
- Cho WC (2010) MicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int J Biochem Cell Biol 42(8): 1273-1281.
- Chang YF, Hung SH, Lee YJ, Chen RC, Su LC, et al. (2011) Discrimination of breast cancer by measuring prostate-specific antigen levels in women’s serum. Anal Chem 83(13): 5324-5328.
- Jemal A, Siegel R, Xu J, Ward E (2010) Cancer statistics. CA Cancer J Clin 60(5): 277-300.
- Siegel RL, Miller KD, Jemal A (2015) Cancer statistics, 2015. CA Cancer J Clin 65(1): 5-29.
- Maillot G, Lacroix-Triki M, Pierredon S, Gratadou L, Schmidt S, et al. (2009) Widespread estrogen-dependent repression of micrornas involved in breast tumor cell growth. Cancer Res 69(21): 8332-8340.
- Ouyang M, Li Y, Ye S, Ma J, Lu L, et al. (2014) MicroRNA profiling implies new markers of chemoresistance of triple-negative breast cancer. PloS One 9(5): e96228.
- Bisso A, Faleschini M, Zampa F, Capaci V, De Santa J, et al. (2013) Oncogenic miR-181a/b affect the DNA damage response in aggressive breast cancer. Cell Cycle 12(11): 1679-1687.
- Ota D, Mimori K, Yokobori T, Iwatsuki M, Kataoka A, et al. (2011) Identification of recurrence-related microRNAs in the bone marrow of breast cancer patients. Int J Oncol 38(4): 955-962.
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, et al. (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 105(30): 10513-10518.
- Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, et al. (2010) The microRNA spectrum in 12 body fluids. Clin Chem 56 (11): 1733-1741.
- Chin LJ, Slack FJ (2008) A truth serum for cancer-microRNAs have major potential as cancer biomarkers. Cell Res 18(10): 983-984.
- Guay C, Regazzi R (2013) Circulating microRNAs as novel biomarkers for diabetes mellitus. Nat Rev Endocrinol 9(9): 513-521.
- Gandhi R, Healy B, Gholipour T, Egorova S, Musallam A, et al. (2013) Circulating microRNAs as biomarkers for disease staging in multiple sclerosis. Ann Neurol 73(6): 729-740.
- Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, et al. (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9(6): 654-659.
- Shen J, Stass SA, Jiang F (2013) MicroRNAs as potential biomarkers in human solid tumors. Cancer Lett 329(2): 125-136.
- Lindner K, Haier J, Wang Z, Watson DI, Hussey DJ, et al. (2015) Circulating microRNAs: emerging biomarkers for diagnosis and prognosis in patients with gastrointestinal cancers. Clin Sci (Lond) 128(1): 1-15.
- Chen X, Ba Y, Ma L, Cai X, Yin Y, et al. (2008) Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 18(10): 997-1006.
- Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, et al. (2008) Serum microRNAs are promising novel biomarkers. PLoS ONE 3(9): e3148.
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, et al. (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 105(30): 10513-10518.
- Chen W, Cai F, Zhang B, Barekati Z, Zhong XY (2013) The level of circulating miRNA-10b and miRNA-373 in detecting lymph node metastasis of breast cancer: Potential biomarkers. Tumor Biol 34(1): 455-462.
- Hamam R, Ali AM, Alsaleh KA, Kassem M, Alfayez M, et al. (2016) MicroRNA expression profiling on individual breast cancer patients identifies novel panel of circulating microRNA for early detection. Sci Rep 6: 25997.
- Sundarbose K, Kartha RV, Subramanian S (2013) MicroRNAs as biomarkers in cancer. Diagnostics 3(1): 84-104.
© 2020. Yi Lu. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






