- Submissions

Full Text
Novel Approaches in Cancer Study
Molecular Detection of Epstein-Barr Virus and Human Cytomegalovirus Antigen Expression in Breast Cancer in Khartoum State, Sudan 2018
Riham E Aidrous1, Abdel Rahim M El Hussein2, Salahaldeen H Dahawi1, Isam M Elkhidir3 and Khalid A Enan2*
1Department of Microbiology, Faculty of Medical Laboratories, Sudan
2Department of Virology, Sudan
3Department of Microbiology and Parasitology, Faculty of Medicine, Sudan
*Corresponding author: Khalid A Enan, Department of Virology, Sudan
Submission: September 03, 2019;Published: September 23, 2019

ISSN:2637-773XVolume3 Issue2
Abstract
Background: Breast cancer is a leading cause of death among women worldwide. The association between Epstein-Barr Virus (EBV) and Human Cytomegalovirus (HCMV) and breast cancer risk still remains controversial making it difficult to determine whether either, both or neither virus is causally associated with breast cancer. The aim of this study was to detect EBV and the expression of the Immediate Early Antigen (IE) of HCMV in breast cancer in Sudanese women.
Method: 60 Formalin-fixed and paraffin-embedded tissue blocks from 42 ductal breast carcinoma and18 benign breast tumors (control group) were obtained from the pathology laboratories of several hospitals in Khartoum State, Sudan, and used in this study. We used PCR to detect EBV and the IE-antigen of HCMV in ductal breast carcinoma and benign breast tumor.
Result: Only 3 (7%) out of 42 ductal breast carcinoma were positive for EBV DNA and none of 18 benign breast tumor (control group) were positive for EBV DNA and all 60 specimens were negative for IE-antigen of HCMV expression.
Conclusion: The results of this study failed to show any relationship between EBV and HCMV and the development of breast carcinoma in the tested specimens.
Keywords: Breast cancer; Polymerase chain reaction; Cytomegalovirus; Epstein-barr virus
Introduction
Breast cancer in women worldwide is considered as the most widespread disease and a most important etiology of mortality [1,2]. Several internal and external factors contribute to the development of this cancer. Internal factors such as age, hormonal effects, lifestyle, obesity, alcohol consumption, smoking, gender, anxiety and stress, genetic predisposition (mutation in BRCA1, 2 and other genes) and family history of breast cancer [3,4]. Exogenous factors include infection with oncogenic viruses such as Mouse Mammary Tumor Virus (MMTV), Human Papilloma Virus (HPV) and Epstein-Barr Virus (EBV). Oncogenic viruses are contributing to 20% of human cancers [5]. Recently, Cytomegalovirus (CMV) has been linked to the development of inflammatory diseases and cancer [6]. EBV It is the causative agent of Infectious Mononucleosis (IM) and has been associated with a growing list of malignancies of both lymphoid and epithelial origin including Burkitt’s lymphoma, B-cell lymphoma in immunocompromised subjects, Hodgkin’s lymphoma, and Nasopharyngeal Carcinoma (NPC). Based on this association, the WHO International Agency for Research on Cancer (IARC) has classified EBV among group I carcinogens which are agents that definitely cause neoplasm in humans [7,8].
EBV can establish a latent status and its genome can persist in the nucleus of infected cells in the form of non-integrated circular episomes with a nucleosomal pattern similar to the host chromatin. Through histone modification and DNA methylation, viral promoters are regulated for the lytic cell cycle and reactivation. The EBV latent episomes have a capacity to influence the epigenetic state of the host DNA and can express several types of oncoproteins which are associated with different type of malignant tumors [9].
A number of mechanisms about the CMV ability in initiating and progressing breast cancer are postulated. Initially, it was revealed that HCMV gene products influence regulation of cell cycle, and inhibit apoptosis; moreover, they activate angiogenesis and metastatic phenotype [10]. Furthermore, HCMV displays immunosuppressive properties, leading to the tumor cells escape from immune surveillance mechanisms [11]. The current study was initiated with the aim of finding out whether EBV and HCMV is associated with breast cancer in Sudanese women.
Material and Method
A Formalin-fixed and paraffin-embedded breast tissue blocks collected during March 2018 to June 2019 from pathology laboratories of different Khartoum hospitals were used. In this study the samples included 42 ductal breast carcinoma and 18 benign breast tumor (Control group).
Deparaffinization
The specimens were deparaffinized using xylene and ethanol (Germany, Merk). Initially, all collected specimens were placed in microtubes; subsequently, xylene was added to them and the tubes were kept at 45 °c for 15 minutes and then centrifuged at 14000RPM. This process was repeated once more. The supernatant was decanted, and 1ml absolute ethanol was added to the precipitate, and kept at room temperature for 10 minutes and was then centrifuged again at 14000RPM for one minute. After casting off the supernatant, 1ml of 70% ethanol was added and the above step was repeated. After this step, the supernatant was discarded again and the microtubes were incubated at 65 °c for five minutes and then the sediments were used for DNA extraction.
DNA extraction
DNA extracted from deparaffinizd tissue by guanidine chloride (Black-well laboratory Cambridge, UK) as fallow: samples were subject to lyses solution containing 400mMNaCl, 6M guanidine chloride and 300μl of 7.5% ammonium acetate and heat treated at 98 °C for 20 minutes in water bath. After cooling10μl (20mg/ml stock) proteinase K was added and incubated for overnight at 56 °C. On day two, second heat treatment was applied by incubating samples at 98 °C for 5 minutes in a water bath. After cooling 10μl proteinase k was added, briefly vortexed and incubated at 56 °C for overnight. During the whole incubation period samples were put on a shaker at interval for about 30 minutes. Chloroform was then added, the supernatant was collected, and DNA was precipitated by ethanol, dissolved in 100μl TE storage buffer. The purity and quality of the extracted DNA was analyzed based on absorbance of the extracted DNA at 260 and 280nm wavelengths using a spectrophotometer (Nano Drop-1000, Thermo Fisher Scientific, and Wilmington, USA).
DNA amplification
The outer prime sequences (Forward: 5-GGA TCC GCA TGG CAT TCA CGT ATG T-3, reverse: 5-GAA TTC AGT GGA TAA CCT GCG GCG A-3) were selected from a conserved region of the fourth exon of the HCMV Immediate Early (IE) gene, located in the Hind III-X fragment of the AD-16n9 strain [12,13] and primers for EBV sequence were (SL1, 5-GGACCTCAAAGAAGAGGGGG-3 and the reverse primer SL3, 5-GCTCCTGGTCTTCCGCCTCC-3. Optimized PCR reaction for HCMV DNA amplification was performed according to [13]. briefly, 3μl of DNA extract was added to PCR premix (Maxime PCR premix kit (i-Tag)) containing i-Tag TM DNA polymerase, dNTP mixture and reaction buffer. Two ul of primers and 13μl of distilled water were then added to PCR premix. The same protocols used for EBV and HCMV DNA amplification are shown in (Table 1). The PCR was carried out in a total volume of 25μl and the amplified PCR product was detected by agarose gel electrophoresis. The product was visualized by staining with 0.15% Ethidium bromide using UV gel documentation system (INGeNius). The expected size of Immediate Early (IE) gene amplicon was 406bp and EBV gene was 80bp.
Table 1:PCR program.
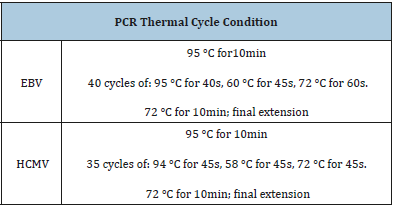
*BC=Breast carcinoma, BBT=Breast Benign Tumor
Statistical analysis
To perform the analysis, Cramer test and SPSS 16 software were used. Statistical significance was set at a P value of less than 0.05.
Ethical Approval
The study was approved by the Ethical Review Committees (ERC) of the Ministry of Health Khartoum State, Sudan and Al Neelain University (ERC), Faculty off Medical Laboratory Sciences.
Result
Table 2:Prevalence of EBV and HCMV in studied groups.
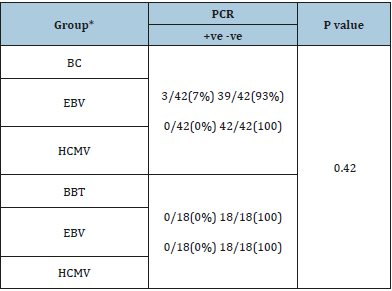
A total of 60 tissue sample were diagnosed. Among the 42 breast carcinoma test specimens, 3 (7%) cases showed positive results for EBV DNA (EBNA) by PCR while none among 18 benign tumor control group was EBV DNA positive but with no significant difference between the two groups (p 0.42) (Table2). The ages of the patients positive for EBV DNA was between 31 to 50 years but no significant differences were discernable among the age groups (p 0.3) (Table3). None of the 60 samples was proved positive for HCMV IE gene.
Table 3:Prevalence of EBV according to age groups.
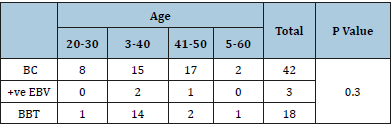
BC=Breast Carcinoma, BBT=Breast Benign Tumor
Discussion
Breasts cancer is a dominant cause of death among women worldwide. The rate of breast cancer is rising about 3% per year [14]. Using PCR-based techniques, a number of studies have reported a positive correlation between EBV and HCMV with breast cancer. The frequency of EBV detection in breast cancer is varied in different region of world. In the present study About 3/42 (7%) of ductal breast carcinoma revealed positive reaction for EBV DNA while none the control group (benign tumor) were positive for EBV DNA but with no significant difference between the two groups (p=0.42). These results indicated that Epstein-Barr virus does not seem to play a significant role in breast cancer in our cohort of women from Khartoum. Our finding is in agreement with Zahra S et al. [15] who showed that only 2 cases out of the 40 tumor samples (5%) were positive and none of 40 normal tissues were positive for EBV-DNA. In addition, Eghbali et al. [16] in Tehran, Iran, reported EBV infection in 16.6% of carcinoma and 4.1% of fibroadenoma samples, but this infection rate was statistically insignificant. A low frequency (7.3%) of EBV DNA have also been reported among the breast cancer in another study in Iran [17].
In contrast, a high frequency of 55.5% of EBV DNA detection in women with breast cancer have been reported in Sudan [18]. In addition, Abdel-Rahman et al. [19], reported EBV DNA in 14/50 (28%) of Iraqi women with breast cancer. Moreover, study by Ghimja F et al. [20] from Eritrea indicated the presence of EBV genome in 27.77% (n=144) of breast cancer samples. Also, in our study the HCMV IE antigen expression was not detected in any of the cases and control group indicating no association between HCMV infection and ductal breast carcinoma. Similar negative results were reported by Mohammadizadeh et al. [21] from Iran who investigated HCMV IE protein expression in 70 breast cancer and 26 normal breast tissues samples using the IHC method and real-time PCR. Another study by Utrera Barillas et al. [22] in Mexico evaluated 27 breast cancer specimens and 20 fibroadenoma samples by quantitative PCR and reported no significant association between HCMV and breast cancer development. Likewise, Antonsson et al. [23] also failed to detect CMV in breast cancer specimens with the quantitative PCR method in 54 Australian breast cancer tumor specimens.
On the other hand, several studies succeeded to show relationship between HCMV and breast cancer. For example, Harkins et al. [24] in the United States demonstrated a higher expression of HCMV antigens in breast cancer compared to normal breast epithelium (97% versus 63%), and Karimi et al. [25] also detected HCMV DNA in 26 of the 50 samples (58%) of invasive breast carcinoma by using the nested-PCR method in Iran. The differed prevalence of EBV and HCMV in various geographical areas may justify the different findings of studies Moreover, the different fixation times of tissue samples in various centers,, and time of exposure to the virus in life (early or late) may also justify some differences in the research findings. Other possibilities for these contrasts, are the limitation of molecular methods used and an absence of virus after carcinogenesis, a so-called “hit and run” oncogenesis [26]. In conclusion our results failed to show any relationship between HBV and HCMV with breast cancer development. Due to the controversial findings concerning the relationship between HBV and HCMV and breast cancer development in several studies in different countries, further studies in this field in our country are mandatory.
References
- Key TJ, Verkasalo PK, Banks E (2001) Epidemiology of breast cancer. Lancet Oncol 2(3): 133-140.
- Taher C, Boniface J, Mohammad AA, Religa P, Hartman J, et al. (2013) High prevalence of human cytomegalovirus proteins and nucleic acids in primary breast cancer and metastatic sentinel lymph nodes. PLoS One 8(2): e56795.
- Parkin DM (2006) The global health burden of infection-associated cancers in the year 2002. Int J Cancer 118(12): 3030-3044.
- Hoffman GL, Clarke JN (2000) Quality of breast cancer sites on the world wide web. Can J Public Health 91(4): 281-284.
- Lawson JS, Heng B (2010) Viruses and breast cancer. Cancers (Basel) 2(2): 752-772.
- Söderberg NC (2006) Does cytomegalovirus play a causative role in the development of various inflammatory diseases and cancer? J Int Med 259(3): 219-246.
- Glaser SL, Hsu JL, Gulley ML (2004) Epstein-barr virus and breast cancer: State of the evidence for viral carcinogenesis. Cancer Epidemiol Biomarkers Prev 13(5): 688-697.
- Ribeiro SA, Ramalho LN, Garcia SB, Zucoloto S (2004) Does the correlation between EBNA-1 and p63 expression in breast carcinomas provide a clue to tumorigenesis in epstein-barr virus-related breast malignancies. Braz J Med Biol Res 37(1): 89-95.
- Ok CY, Li L, Young KH (2015) EBV-driven B-cell lymphoproliferative disorders: from biology, classification and differential diagnosis to clinical management. Exp Mol Med 47: e132.
- Dziurzynski K, Chang SM, Heimberger AB, Kalejta RF, Dallas SRM, et al. (2012) Consensus on the role of human cytomegalovirus in glioblastoma. Neuro Oncol 14(3): 246-55.
- Mohammed AH, Kadhim HS, Hussein AA (2015) Investigation the role of human cytomegalovirus in the invasive ductal breast carcinoma using real time PCR. Int J Curr Microbiol App Sci 4(1): 537-542.
- Drouet E, Michelson S, Denoyel G, Colimon R (1993) Polymerase chain reaction detection of human cytomegalovirus in over 2000 blood specimens correlated with virus isolation and related to urinary virus excretion. J Virol Methods 45(3): 259-276.
- Mendez JC, Espy MJ, Smith TF, Wilson JA, Paya CV (1998) Evaluation of PCR primers for early diagnosis of cytomegalovirus infection following liver transplantation. J Clin Microbiol 36(2): 526-530.
- Forouzanfar MH, Foreman KJ, Delossantos AM, Lozano R, Lopez AD, et al. (2011) Breast and cervical cancer in 187 countries between 1980 and 2010: A systematic analysis. Lancet 378(9801): 1461-1484.
- Zahra S, Faranak Hadi, Seyed HH, Zahra S (2018) The relationship between EBV virus and breast cancer in Khuzestan province of Iran. J Appl Biotechnol Re 5(1): 37-41.
- Eghbali M, Ghane M, Mirinargesi M, Zare MA (2012) Detection of epstein barr virus among benign and malignant breast tumors. Asian J Exp Biol Sci 5(7): 365-371.
- Torabizadeh Z, Nadji A, Naghshvar F, Anahita N, Mohsen P (2014) Association between epstein-barr virus (EBV) and breast cancer. Res Mol Med 2(4): 24-29.
- Yahia ZA, Adam AA, Elgizouli M, Hussein A, Masri MA, et al. (2017) Epstein barr virus: A prime candidate of breast cancer etiology in Sudanese patients. Infect Agent and Cancer 9(1): 9.
- Abdel RNZ, Abeer AB, Waleed S, Kassem FA, Khalidi SJ, et al. (2012) Epstein-barr virus and breast cancer: Epidemiological and molecular study on Egyptian and Iraqi women. J Egypt Natl Canc Inst 24(3): 123-131.
- Ghimja F, Ahmed ME, Elwaleed ME, Ameera AMA, Anghesom G, et al. (2017) Association of epstein-barr virus and breast cancer in Eritrea. Infectious Agents and Cancer 12: 62.
- Mohammadaizadeh, Fatemeh M (2017) Evaluation of human cytomegalovirus antigen expression in invasive breast carcinoma in a population of Iranian patient. Infectious Agents and Cancer 12: 39.
- Utrera BD, Valdez SHA, Gómez RD, Alvarado CI, Aguilera P, et al. (2013) Is human cytomegalovirus associated with breast cancer progression. Infectious agents and cancer 8(1): 12.
- Antonsson A, Bialasiewicz S, Rockett RJ, Jacob K, Bennett IC, et al. (2012) Exploring the prevalence of ten polyomaviruses and two herpes viruses in breast cancer. PLoS ONE 7(8): e39842.
- Harkins LE, Matlaf LA, Soroceanu L, Klemm K, Britt WJ, et al. (2010) Detection of human cytomegalovirus in normal and neoplastic breast epithelium. Herpesviridae 1(1): 8.
- Karimi M, Hosseini SZ, Nikkhoo B, Soleimani MF (2016) Relative frequency of Cytomegalovirus (CMV) in tissue samples of women with breast cancer in Sanandaj, Iran. International Journal of Bioassays 5(3): 4907-4911.
- Richardson AK, Currie MJ, Robinson BA, Morrin H, Phung Y, et al. (2015) Cytomegalovirus and epstein-barr virus in breast cancer. PLoS One 10(2): e0118989.
© 2019 Khalid A Enan. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






