- Submissions

Full Text
Modern Research in Dentistry
Expression of the Programmed Death Ligand (PDL-1) CD274 in Oral Squamous Cell Carcinomas
Carlos Atrian, Jorge Quiroz, Dana Thimons and Karl Kingsley*
School of Dental Medicine, University of Nevada, USA
*Corresponding author: Karl Kingsley, Professor of Biomedical Sciences and Director of Student Research, University of Nevada, Las Vegas-School of Dental Medicine 1001 Shadow Lane, B313 Las Vegas, Nevada 89106, USA
Submission: February 23, 2018; Published: March 14, 2018

ISSN:2637-7764Volume2 Issue1
Abstract
Background: The programmed death ligand or PDL-1 (also known as B7-H1) has been shown to inhibit tumor immunology and facilitate immune evasion. Although these mechanisms are well known in many cancers, recent evidence has suggested this function may also be active in oral cancers. To determine appropriate models to study this phenomenon, well-characterized oral cancer cell lines were screened to evaluate the presence and expression of PDL-1 or B7-H1.
Methods: Commercially available oral squamous cell carcinomas (SCC25, SCC15, CAL27) were used for this study. RNA was isolated and subsequently screened using validated primers specific for B7-H1/PDL-1. RNA was quantified and screened for purity prior to normalization and the RT-PCR screening procedure.
Results: mRNA was successfully isolated from each cell line and verified using spectrophotometric measurements and internal PCR control standards. Subsequent screening using validated primer sets revealed expression of B7-H1 in all oral cancer cell lines, with differential expression observed using the PDL-1 validated primer set. No expression was observed in the normal, non-cancerous oral gingival cell line examined.
Conclusion: These results demonstrated that the oral cancer cell lines evaluated expression mRNA which can be successfully screened using previously validated primers. However, because differential non-equivalent results were obtained when comparing the two validated primer sets, care should be taken to determine why the difference in specificity was observed and how this may affect selection and evaluation of appropriate screening tests for future clinical samples and isolates.
Background
B7-H1 (also known as programmed death ligand or PDL-1) is an inhibitory molecule of T cells implicated in the protection of tumor cells from immune targeting [1,2]. Recent evidence has suggested some proportion of squamous cell carcinomas of the head and neck (SCCHN) expressed B7-H1, which has also been reported in breast, cervical, esophageal and colorectal cancers [3,4]. B7/PDL-1 binds the cognate receptor programmed death (PD-1) on cytotoxic T-cells, which leads to apoptosis and thus protection from lysis by activated T-cell (CTL) surveillance [5,6].
Although several studies have reported screening for B7-H1 or PDL-1 among clinical patient samples, few if any have evaluated whether this expression is found among well-characterized oral cancer cell lines [7,8]. Many of these cell lines are oral squamous cell carcinomas and are therefore appropriate models to study the potential for therapeutic targets to modulate expression of this cell surface ligand. Due to the ability of this ligand to down-regulate the activity of CD8 or cytotoxic T-cells, this pathway inhibits CD8- specitic targeting of mutated oncoproteins expressed on Class I major histocompatibility or MHC I molecules thereby allowing immune evasion [9-11].
Based upon the importance of this function in tumor immunity, the primary goal of this study was to evaluate commercially available oral cancer cell lines to determine if B7-H1/PDL-1 mRNA can be detected. If successful, this would provide the foundation for future studies to develop and test targeted therapies to inhibit or impair the expression and function of this ligand in oral cancers.
Methods
Cell culture
Commercially available cell lines were obtained from American Tissue Culture Collection (AATCC), Manassas, VA. Several oral squamous cell carcinoma cell lines were obtained (SCC25, SCC15 and CAL27), as well as a normal human gingival fibroblast cell line (HGF-1). Cells were cultured according to the manufacturer protocol, which included Dulbecco's Modified Eagle's Medium (DMEM) with 4.0mML-Glutamine -adjusted to also contain 3.7g/L sodium bicarbonate, 4.5g/L glucose and 110mg/L sodium pyruvate. All cell cultures were supplemented with Penicillin (10,000units/ mL) and Streptomycin (10,000ug/mL) with additional 10% fetal bovine serum (FBS) from HyClone (Logan, UT). Cells were maintained and all experiments performed in a BSL-2 approved, humidified cell culture incubator at 37 °C and 5% CO2 as previously described [12,13].
RNA isolation
RNA was isolated from confluent cell cultures in 25cm2 BD Falcon tissue-culture treated flasks. Cell numbers approximated 1.2 -3.5 x 107 cells. RNA extraction was performed using the AB gene Total RNA isolation reagent kit from Epson (Surrey, UK) and the protocol recommended by the manufacturer. RNA purity and quantity were determined using spectrophotometer absorbance readings from each isolate at 260 and 280nm.
More specifically, diluted RNA isolates from each cell line (10uL of RNA extraction in 490uL nuclease-free water, pH 7.0) was used for analysis. RNA purity can then be determined from the ratio of A260:A280 absorbance, which should be approximately 1.80 or higher. RNA concentrations were then calculated using the formula:
40 x A260 (Absorbance reading) x 50 (dilution factor) = sample yield per mL
PCR standards
To ensure all reverse-transcription polymerase chain screening reactions (RT-PCR) were processed properly, structural and biochemical mRNA standards were used to screen each RNA sample isolate. Primers for the structural cytoskeletal component beta actin were obtained, as well as for the glycolytic pathway enzymeglyceraldehyde-3-phosphate dehydrogenase (GAPDH). Equal amounts of RNA (1ng) from each isolate were then used to screen for mRNA expression of beta actin and GAPDH and all samples with positive expression were available for the current screening study.
RT-PCR screening
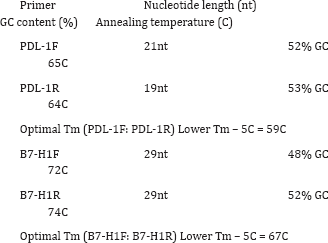
Each RNA sample isolate was then processed using primers for B7-H1 / PDL-1 and the ABgene Reverse-iT One-Step RT-PCR kit and protocol. In brief, reverse transcription at 47 °C was performed for 30 minutes, which was then followed by 35PCR amplification cycles -including 20 seconds of denaturation, 30 seconds of annealing at the optimal primer set temperature with five minutes of final extension at 72 °C (PDL-1 59C; B7-H1 67C). PCR products were visualized using gel electrophoresis and imaged using a Kodak Gel Logic 100 Imaging System (Rochester, NY).
PDL-1F: GGT GAG GAT GGT TCT ACA CAG (21 nt, 52% GC, Tm 65C)
PDL-1R: GAG AAC TGC ATG AGG TTG C (19 nt, 53% GC, Tm 64C)
Optimal Tm=59C
B7-H1F: AAA GGT ACC TAG AAG TTC AGC GCG GGA TA
B7-H1R AAA GGA TCC CAG CGA GCT AGC CAG AGA TA
Results
Table 1: Quantification of RNA isolation concentration and purity.
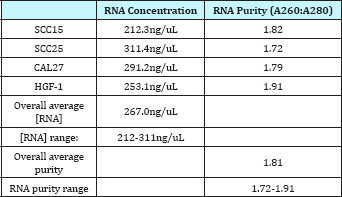
RNA isolation from each cell line was successful, with an overall average concentration of257ng/uL, which was well within the range specified by the manufacturer of 100 -1000ng/uL (Table 1). The overall average RNA purity measured by A260:A280 absorbance readings was 1.81, which is considered pure (A260:A280 >1.80), which ranged from 1.72 to 1.91. These data provided verification and quantification of the RNA isolated from each cell line sample.
To ensure the requisite quality and integrity of all reverse- transcription polymerase chain screening reactions (RT-PCR), structural and biochemical mRNA standards were then utilized to screen each RNA sample isolate (Figure 1). This screening revealed strong, consistent expression of mRNA specific for the structural protein-actin among all cell lines, as well as detectable expression of mRNA for the glycolytic pathway enzymeglyceraldehyde-3- phosphate dehydrogenase (GAPDH). These results also revealed differential results for GAPDH expression with the strongest expression observed among the most rapidly growing oral cancers (SCC25, CAL27).
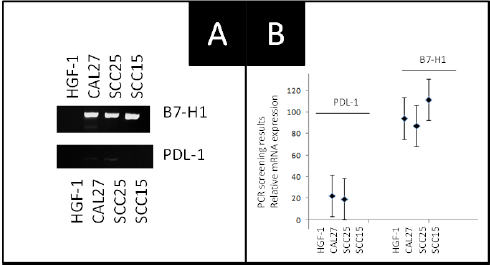
Based upon these findings, RNA from all samples was then screened for B7-H1 or PDL-1 mRNA using two distinct primer sets (Figure 2). The results of this screening demonstrated that the oral cancer cell lines (CAL27, SCC15, SCC25) expressed B7-H1 mRNA, although the normal human gingival fibroblast did not. In addition, the second validated primer set for PDL-1 yielded weakly positive results with both CAL27 and SCC15 - although no results were observed with SCC15 or the normal HGF-1 cell line.
Figure 1: RT-PCR screening of cell line isolates using PCR standards. A) RT-PCR for mRNA using structural (b-actin) and metabolic (GAPDH) proteins revealed expression in all cell lines. B) GAPDH (metabolic) expression was strongest among the most rapidly proliferating cells (CAL27, SCC25) as determined by confluence at three days (3d.) following a 1:4 split.
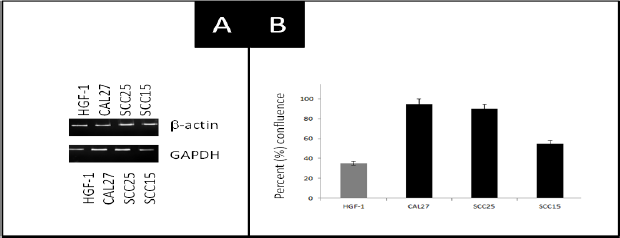
Figure 2: RT-PCR screening of RNA extractions from all cell lines reveals differential results. A) The validated primer sets for B7-H1 and PDL-1 mRNA revealed differential expression in CAL27, SCC15 and SCC25 but not HGF-1 cells. Positive results were observed in SCC25 and CAL27 cells with B7-H1 but not PDL-1. B) Analysis of RT-PCR band intensity revealed strong specific bands using the B7-H1 validated primers in all oral cancer lines but weakly positive results in CAL27 and SCC25 using PDL-1.
Discussion
The primary goal of this study was to evaluate commercially available oral cancer cell lines to determine if B7-H1 / PDL-1 mRNA can be detected. The results of study demonstrated that the oral squamous cell carcinomas evaluated had sufficient mRNA for detection, which had greater specificity and stronger band intensity using the B7-H1 primer set compared with the PDL-1 primer sequences. In addition, although positive results were obtained in both experiments, differential results were observed with PDL-1, which may suggest the B7-H1 primers would be the most appropriate screening tool for evaluating this specific biomarker among clinical samples.
The results of the structural (beta actin) and metabolic (GAPDH) mRNA analysis provide additional support and evidence that the RNA isolation, yield and purity were sufficient to facilitate this type of analysis. Furthermore, the RT-PCR band intensity also revealed specific and quantifiable results that clearly demonstrated the sensitivity of the screening protocol and PCR assays.
Although these results clearly demonstrated that some oral squamous cell carcinomas express mRNA specific for the PDL-1 / B7-H1 ligand, some limitations were inherent to this study which must also be discussed. For example, only a limited subset of oral cancer cell lines were available for this screening - which could be expanded upon in future studies of this nature. In addition, no primary tumor isolates were available to the study authors - which would provide further evidence of the screening potential for this type of biomarker assay.
Despite these limitations, these results may be among the first to clearly demonstrate strong and specific expression of the PDL-1/ B7-H1 ligand in oral cancers. These data suggest that further studies are warranted to determine if this observation can be made in all other commercially available oral cancers before screening studies can begin on primary tumor isolates or other types of clinical samples. However, these results strongly suggest more research is needed due to the importance of this ligand in immunologic evasion [14,15].
a. Choose the appropriate size of the bite tray
b. Apply material on both sides
c. Reasonable material thickness (4 to 5mm)
d. Elimination of material in respect to the presumed treatment areas while trying to preserve tow points of contact on each side or at least one point on each side.
e. This technique is still under technical development and needs more clinical trials to achieve its most complete specifications.
Some modifications might be implemented on the used materials (trays) simplifying the data reading and better understanding the brain interpretation to these stimulating signals.
References
- Alsaab HO, Sau S, Alzhrani R, Tatiparti K, Bhise K, et al. (2017) PD-1 and PD-L1 checkpoint signaling inhibition for cancer immunotherapy: mechanism, combinations, and clinical outcome. Front Pharmacol 8: 561.
- Diesendruck Y, Benhar I (2017) Novel immune check point inhibiting antibodies in cancer therapy-opportunities and challenges. Drug Resist Updat 30: 39-47.
- Mishra A (2017) PD-1/PD-L1 biology and immunotherapy in HPV- positive oral cancers. Future Oncol doi: 10.2217/fon-2017-0115.
- Hoesli R, Birkeland AC, Rosko AJ, Issa M, Chow KL, et al. (2018) Proportion of CD4 and CD8 tumor infiltrating lymphocytes predicts survival in persistent/recurrent laryngeal squamous cell carcinoma. Oral Oncol 77: 83-89.
- Heymann MF, Lezot F, Heymann D (2017) The contribution of immune infiltrates and the local microenvironment in the pathogenesis of osteosarcoma. Cell Immunol pii: S0008-8749(17)30189-2.
- Spurrell EL, Lockley M (2014) Adaptive immunity in cancer immunology and therapeutics. Ecancermedicalscience 8: 441.
- Hirai M, Kitahara H, Kobayashi Y, Kato K, Bou-Gharios G, et al. (2017) Regulation of PD-L1 expression in a high-grade invasive human oral squamous cell carcinoma microenvironment. Int J Oncol 50(1): 41-48.
- Lanzel EA, Paula Gomez Hernandez M, Bates AM, Treinen CN, Starman EE, et al. (2016) Predicting PD-L1 expression on human cancer cells using next-generation sequencing information in computational simulation models. Cancer Immunol Immunother 65(12): 1511-1522.
- Wu L, Deng WW, Yu GT, Mao L, Bu LL, et al. (2016) B7-H4 expression indicates poor prognosis of oral squamous cell carcinoma. Cancer Immunol Immunother 65(9): 1035-1045.
- Moore EC, Cash HA, Caruso AM, Uppaluri R, Hodge JW, et al. (2016) Enhanced tumor control with combination mTOR and PD-L1 inhibition in syngeneic oral cavity cancers. Cancer Immunol Res 4(7): 611-620.
- Yu GT, Bu LL, Huang CF, Zhang WF, Chen WJ, et al. (2015) PD-1 blockade attenuates immunosuppressive myeloid cells due to inhibition of CD47/ SIRPa axis in HPV negative head and neck squamous cell carcinoma. Oncotarget 6(39): 42067-42080.
- Osafi J, Hejazi A, Stutz DD, Keiserman MA, Bergman CJ, et al. (2014) Differential effects of 1,25-dihydroxyvitamin D3 on oral squamous cell carcinomas in vitro. J Diet Suppl 11(2): 145-154.
- Moody M, Le O, Rickert M, Manuele J, Chang S, et al. (2012) Folic acid supplementation increases survival and modulates high risk HPV- induced phenotypes in oral squamous cell carcinoma cells and correlates with p53 mRNA transcriptional down-regulation. Cancer Cell Int 12: 10.
- Tsushima F, Tanaka K, Otsuki N, Youngnak P, Iwai H, et al. (2006) Predominant expression of B7-H1 and its immunoregulatory roles in oral squamous cell carcinoma. Oral Oncol 42(3): 268-274.
- Concha-Benavente F, Srivastava RM, Trivedi S, Lei Y, Chandran U, et al. (2016) Identification of the cell-intrinsic and -extrinsic pathways downstream of EGFR and IFNy that induce PD-L1 expression in head and neck cancer. Cancer Res 76(5): 1031-1043.
© 2018 Carlos Atrian, et al. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






