- Submissions

Full Text
Modern Concepts & Developments in Agronomy
The Most Popular Measures of Genetic Similarity Based on Analyses of DNA Polymorphism
Jan Bocianowski*
Department of Mathematical and Statistical Methods, Poznań University of Life Sciences, Poland
*Corresponding author: Jan Bocianowski, Department of Mathematical and Statistical Methods, Poznań University of Life Sciences, Poland Add: ORCID: 0000-0002-0102-0084
Submission: May 11, 2020Published: May 19, 2020

ISSN 2637-7659Volume 6 Issue 3
Opinion
Genetic similarity of genotypes is the very important for analysis of quantitative traits in all organisms. All organisms are exposed on the influence of different environmental conditions but in the genetic level are the same in all environments. A number of studies have shown that the greater the genetic similarity of parental lines, the smaller the heterosis effect [1]. Genetic or phenotypic similarity can be estimated by genotype testing on the basis of the observations obtained through prediction (a priori) or of the observations and studies (a posteriori). Coors [2] stated that predicting the effect of heterosis between groups of germplasm showing genetic similarity of germplasm was not possible on the basis of the genetic distance determined with using the DNA markers, but should be determined in the field experiments. The paper presents five the most popular methods of estimation of genetic similarity (S) based on coefficients proposed by Jaccard, Kulczynski, Sokal and Michener, Nei as well as Rogers.
Jaccard [3]
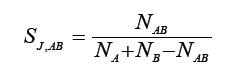
where NA-number of bands present in genotype A, NB-number of bands present in genotype B, NAB-number of bands present in genotypes A and B.
Kulczyński [4]
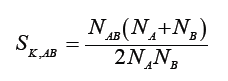
Sokal & Michener [5]
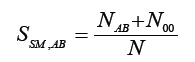
where N00 number of lack of bands present in both, A and B, genotypes, N-number of all markers.
Nei [6]
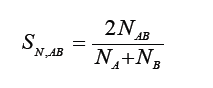
Rogers [7]
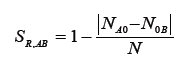
Where NA0 number of band present in genotype A and absent in genotype B, N0B-number of band present in genotype B and absent in genotype A.
References
- Tomkowiak A, Bocianowski J, Kwiatek M, Kowalczewski PŁ (2020) Dependence of the heterosis effect on genetic distance, determined using various molecular markers. Open Life Sciences 15(1): 1-11.
- Coors JG, Pandey S, Melchinger AE (1999) Genetic diversity and heterosis. In: The Genetics and Exploitation of Heterosis in Crops. American Society of Agronomy, Crop Science Society of America, Wisconsin, USA, pp. 99-118.
- Jaccard P (1908) Nouvelles recherches sur la distribution florale. Bull la Société vaudoise des Sci Nat 44: 223-270.
- Kulczyński S (1927) Die Pflanzenassoziationen der Pieninen. Bull Int L Académie Pol des Sci des Lett Cl des Sci Math Nat 2: 57-203.
- Sokal RR, Michener CD, Sokal R, Michener C, Sokal RR, et al. (1958) A statistical method for evaluating systematic relationships. Univ Kansas Sci Bull 38: 1409-1438.
- Nei M (1972) Genetic distance between populations. The American Naturalist 106(949): 283-292.
- Rogers JS (1972) Measures of genetic similarity and genetic distance. In: Studies in Genetics VII. University of Texas Publication, Austin, TX, USA, pp. 145-153.
© 2020 Jan Bocianowski. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






