- Submissions

Full Text
Journal of Biotechnology & Bioresearch
Yeast Autophagy and Life-Span Regulation: A Mini-Review
Jinru Yang1, Wen Li1, Huabiao Miao1,2,3 and Zunxi Huang1,2,3*
1School of Life Sciences, Yunnan Normal University, Kunming, Yunnan 650500, China
2Engineering Research Center of Sustainable Development and Utilization of Biomass Energy, Ministry of Education, Kunming, Yunnan 650500, China
3Key Laboratory of Yunnan Province for Biomass Energy and Biotechnology of Environment, Kunming, Yunnan 650500, China
*Corresponding author:Zunxi Huang, School of Life Sciences, Yunnan Normal University, Engineering Research Center of Sustainable Development and Utilization of Biomass Energy, China
Submission: July 11, 2025;Published: July 28, 2025

Volume5 Issue 5July 28, 2025
Abstract
From the pursuit of eternal life in ancient civilizations to the comprehensive study of aging mechanisms in modern science, humanity has consistently focused on the phenomenon of aging. In aging research, yeast is frequently utilized as a model organism due to its straightforward genetic manipulation and conserved aging pathways. Recent studies increasingly demonstrate that autophagy plays a crucial role in regulating cellular lifespan and aging, with its mechanisms involving oxidative stress, genomic stability, and metabolic regulation. This review primarily examines the impact of autophagy on the regulation of yeast cell lifespan and explores how modulating autophagy can delay cellular aging, offering insights that may be applicable to other organisms in their quest to postpone aging.
Keywords:Aging; Yeast; Autophagy; Life-span regulation
Introduction
Aging is a complex biological process, involving the interaction of heredity, cell molecules, environment, lifestyle and other aspects [1]; And aging often leads to the irreversible decline of body function with the increase of age, which leads to a variety of pathologies and diseases, such as cardiovascular diseases, neurodegeneration and cancer [2,3], which brings huge economic burden to society and families [4]. Further approaches that are able to regress (or at least delay) the onset of pathogenic age-related decline are urgently needed. In recent years, such studies have been performed in a wide array of model organisms, from in vivo vertebrate and invertebrate animal studies to in vitro donor-derived cultured cell studies, as well as experiments in the highly genetically tractable yeast [5]. Strikingly, many of the mechanisms that contribute to cellular aging appear to be conserved from yeast to humans [6]. And some studies found that autophagy, oxidative stress, and telomerase activity are key mechanisms related to aging. At the same time, a study found that ATG may act through multiple mechanisms to become an effective anti-aging molecule [4]. Therefore, the study of aging mechanism mostly focuses on the study of autophagy mechanism, and through the regulation of autophagy to achieve the treatment of many chronic diseases and delay the life of the body. Autophagy is a highly conserved subcellular catabolic pathway in eukaryotes. It is a process in which cells break down damaged protein, dysfunctional mitochondria and even intracellular invasive microorganisms through lysosomes or vacuoles to provide energy for cells, maintain intracellular physiological balance and help cells survive adversity. It is very important for development, cell survival and degradation of dysfunctional organelles and potentially toxic aggregates [7], and it is a key regulator for maintaining cell stability [8]. It plays an important role in prolonging life, and it has been shown that increasing autophagy can delay aging [9-14]. Therefore, in this review, we focus on some molecular mechanisms of autophagy and longevity regulation and how to delay the aging of organisms by regulating autophagy.
Core molecular mechanism of autophagy
Types of yeast autophagy: Under normal physiological conditions, autophagy in the organism is maintained at a relatively low level, and it plays the role of cell housekeeper to maintain the intracellular environmental homeostasis; However, under the pressure stimulation of long-term hunger, nutritional deficiency, tissue development and reconstruction, oxidative stress and organelle damage, the body can induce autophagy and degrade its own components, providing a minimum source of nutrition for survival. Autophagy mainly decomposes damaged protein, dysfunctional mitochondria and even intracellular invasive microorganisms through lysosomes or vacuoles in cells. Therefore, autophagy can be divided into macroautophagy, microautophagy, chaperone-mediated autophagy, and DNautophagy/RNautophagy [15] according to the different ways in which substrates enter lysosomes. Among them, macro autophagy accounts for most autophagy activity, which is the main degradation pathway for accumulating protein and damaged organelles in cells. In addition, according to the specificity of autophagy degradation substrate, yeast autophagy can be divided into selective autophagy and non-selective autophagy [13]; Among them, according to the different types of substrates, selective autophagy can be divided into mitophagy, ribophagy, reticulophagy/ ER-phagy, pexophagy, lysophagy, lipophagy, nucleophagy, xenophagy [14].
Figure 1:Mechanism of autophagy [12].
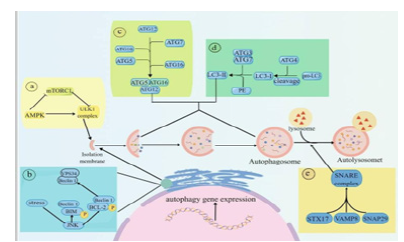
Core molecular mechanism of autophagy: Autophagy is a complex process of self-degradation. The current research shows that autophagy involves five key steps: (1) Induction and regulation of autophagy: the induction of autophagy is regulated by many ways, among which AMPK and mTOR signal transduction pathways can start autophagy, and their functions are to regulate the opening of autophagy through the response of cells to stress (such as hunger or oxidation); (2) Autophagy precursor assembly: activated kinase JNK destroys Beclin1/BCL-2 and Beclin1/BIM complexes by phosphorylating bcl-2 and BIM to release Beclin1; Free Beclin1 activates VPS34 and combines with it to form complex PI3K, which produces PI3P to promote the extension of autophagic vesicles [15]. (3) Extension of autophagy: PI3P produced by PI3P complex can recruit ATG proteins, and these recruited proteins form complex ATG5-ATG12-ATG16L, and then these complexes fuse with autophagy vesicles to form autophagy. (4) LC3 is inserted into autophagy through a series of reactions. (5) Autophagy and lysosome fuse to form autophagy lysosome. STX17 combines with SNAP29 and VAMP8 to form SNARE complex, which is transferred to autophagy membrane, so that lysosomes fuse with autophagy and form autophagy lysosomes. Finally, cell goods are degraded into small molecules by lysosomes and recycled (Figure 1).
Autophagy and yeast longevity regulation: There are two commonly used methods to measure yeast cell senescence: sequential life (CLS) or replication life (RLS) [1,16]. The measurement of replication lifetime (RLS) is to calculate the number of daughter cells produced by each actively dividing mother cell. The sequential life (CLS) is measured by the best ability of culture in quiescent period (G0) to survive for a period of time [16,17].
Moderately enhance autophagy and delay aging: A large number of studies have found that increasing autophagy in moderation can delay aging. For example, the overexpression of Sir2 can significantly prolong the RLS of yeast replication aging, and it is accompanied by the up-regulation of autophagy level [1]; Siqi Chen [18] and others found that ATG can enhance autophagy by targeting protein phosphatase 2A (PP2A), thus prolonging the life span. At the same time, the antioxidant capacity of ATG was evaluated, and it was found that ATG increased the activity of antioxidant enzymes, thus reducing the levels of reactive oxygen species (ROS) and malondialdehyde (MDA), thus improving the survival rate of yeast under oxidative stress. In addition, ATG can improve the expression of EST1, EST2 and EST3 genes in yeast. Hung Ching Cui et al. found that deletion of autophagy-related gene ATG8 or SAC1 shortened the replication life of yeast cells.
Cell death caused by imbalance of autophagy regulation: Autophagy is the main means for cells to degrade the whole organelle, but this self-degradation ability of cells is very risky. Proper autophagy can delay the life of cells, but excessive autophagy is likely to be harmful to organisms. Some recent studies have found that autophagy can also occur in the nucleus, which is called macronuclear autophagy and micronucleus autophagy [19-21]. In the absence of nitrogen, abnormal enhancement of micronucleus autophagy will occur in the nucleus of ATG39- deficient mutant yeast strains, leading to excessive degradation of nuclear components and accelerating cell death [22]. In addition, excessive endoplasmic reticulum autophagy can also cause cell death. Endoplasmic reticulum is the largest organelle in eukaryotic cells, which can form a complex cell quality control network by contacting with other organelle membranes, and participate in Ca2+ homeostasis and protein synthesis [23]. In the process of excessive endoplasmic reticulum autophagy, autophagic vacuoles produced are about five times that produced in cells under physiological conditions and twice that produced under stress [24]. Double autophagy means double energy consumption, and a large amount of energy consumption eventually accelerates cell aging and even leads to cell death [12].
Natural compounds prolong yeast life through autophagy: Natural compounds are chemical substances with pharmacological or biological activities produced by plants, animals and microorganisms in the natural world. Many studies show that natural compounds have certain therapeutic effects on many chronic diseases, and the appearance of these chronic diseases is inseparable from the growth of the body’s age, especially in aging organisms. Autophagy is a means to delay aging, and it seems to be an effective way to use natural compounds to regulate autophagy pathway to delay aging. Many research results also prove the feasibility of this method. In recent years, many studies have reported that plant-derived compounds exert anti-aging effects by inducing autophagy. In the present Yanan Liu study, replicative lifespan and chronological lifespan assays of yeast were used to double-screen antiaging compounds from Gentiana rigescens Franch, a Chinese herb medicine. Inokosterone from G. rigescens Franch extended not only the replicative lifespan of K6001 yeast but also the chronological lifespan of YOM36 yeast. Furthermore, it can enhance the survival ability of mammalian cells [25]. In Siqi Chen study, arctigenin (ATG), a senomorphic, was screened from the Chinese medicine Fructus arctii using K6001 yeast replicative lifespan. Autophagy, oxidative stress, and telomerase activity are key mechanisms related to aging. Furthermore, The Didac Carmona-Gutierrez team research reported the identification of the flavonoid 4,4′-dimethoxychalcone (DMC) as a natural compound with anti- ageing properties. External DMC administration extends the lifespan of yeast, worms and flies, decelerates senescence of human cell cultures, and protects mice from prolonged myocardial ischaemia. Concomitantly, DMC induces autophagy, which is essential for its cytoprotective effects from yeast to mice.
Summary
Here, we briefly review the molecular mechanism related to autophagy pathway and the regulation mechanism of autophagy on cell life, but autophagy is a complex process caused by many factors, and the current research can’t fully explain it, and the diversity of organisms also determines the multifaceted nature of autophagy, and many problems have not been solved so far. In particular, the use of natural compounds to induce autophagy and thus delay aging, in this process, the selection and dosage of natural compounds and how to accurately regulate autophagy to achieve the normal physiological process of cells and the mechanism of delaying cell aging are still unclear, which limits the application of plant-derived compounds in the treatment of many diseases caused by aging..
Acknowledgment
We gratefully acknowledge the financial support by Yunnan Characteristic Plant Extraction Laboratory Co., Ltd., Yunnan 650106, China (YKKF2024007) and Yunnan Province University Service Key Industry Science and Technology Special Project (FWCY-ZNT2024009).
References
- Tyler J, Johnson J (2018) The role of autophagy in the regulation of yeast life span. Ann N Y Acad Sci 1418(1): 31-43.
- Niccoli T, Partridge L (2012) Ageing as a risk factor for disease. Curr Biol 22(17): R741-R752.
- Hou Y, Dan X, Babbar M, Wei Y, Hasselbalch SG, et al. (2019) Ageing as a risk factor for neurodegenerative disease. Nat Rev Neurol 15(10): 565-581.
- Chen, S, Li Y, Wu E, Li Q, Xiang L, et al, (2024) Arctigenin from Fructus arctii exhibits antiaging effects via autophagy induction, antioxidative stress, and increase in telomerase activity in yeast. Antioxidants (Base l) 13(6): 684.
- Longo V, Fabrizio P (2012) Chronological aging in saccharomyces cerevisiae. Subcellular Biochemistry 57: 101-121.
- McCormick MA, Delaney JR, Tsuchiya M, Tsuchiyama S, Shemorry A, et al. (2015) A comprehensive analysis of replicative lifespan in 4,698 single-gene deletion strains uncovers conserved mechanisms of aging. Cell Metab 22(5): 895-906.
- Delorme-Axford E, Klionsky DJ (2018) Transcriptional and post-transcriptional regulation of autophagy in the yeast Saccharomyces cerevisiae. J Biol Chem 293(15): 5396-5403.
- Levine B, Kroemer G (2019) Biological functions of autophagy genes: A disease perspective. Cell 176(1-2): 11-42.
- Liu Y, Liu Q, Chen D, Matsuura A, Xiang L, et al. (2022) Inokosterone from Gentiana rigescens franch extends the longevity of yeast and mammalian cells via antioxidative stress and mitophagy induction. Antioxidants (Basel) 11(2): 214.
- Glick D, Barth S, Macleod K (2010) Autophagy: Cellular and molecular mechanisms. J Pathol 221(1): 3-12.
- Parzych K, Klionsky D (2014) An overview of autophagy: Morphology, mechanism, and regulation. Antioxid Redox Signal 20(3): 460-473.
- Liu S, Yao S, Yang H, Liu S, Wang Y, et al. (2023) Autophagy: Regulator of cell death. Cell Death Dis 14(10): 648.
- Wilfling F, Lee CW, Erdmann PS, Zheng Y, Sherpa D, et al, (2020) A selective autophagy pathway for phase-separated endocytic protein deposits. Mol Cell 80(5): 764-778.e7.
- Kraft C, Peter M, Hofmann K (2010) Selective autophagy: Ubiquitin-mediated recognition and beyond. Nat Cell Biol 12(9): 836-841.
- Hase K, Raluca Contu VR, Kabuta C, Sakai R, Takahashi M, et al. (2020) Cytosolic domain of SIDT2 carries an arginine-rich motif that binds to RNA/DNA and is important for the direct transport of nucleic acids into lysosomes. Autophagy 16(11): 1974-1988.
- Nascimbeni A, Codogno P, Morel E (2017) Phosphatidylinositol-3-phosphate in the regulation of autophagy membrane dynamics. FEBS J 284(9): 1267-1278.
- Fabrizio P, Hoon S, Shamalnasab M, Galbani A, Wei M, et al, (2010) Genome-wide screen in Saccharomyces cerevisiae identifies vacuolar protein sorting, autophagy, biosynthetic, and tRNA methylation genes involved in life span regulation. PLoS Genet 6(7): e1001024.
- Cui H, Cui X, Yang X, Cui X, Sun Y, et al. (2024) Effect of ATG8 or SAC1 deficiency on the cell proliferation and lifespan of the long-lived PMT1 deficiency yeast cells. FEMS Microbiol Lett 371.
- Mochida K, Oikawa Y, Kimura Y, Kirisako H, Hirano H, et al. (2015) Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature 522(7556): 359-362.
- Tomioka Y, Kotani T, Kirisako H, Oikawa Y, Kimura Y, et al. (2020) TORC1 inactivation stimulates autophagy of nucleoporin and nuclear pore complexes. J Cell Biol 219(7): e201910063.
- Lee CW, Wilfling F, Ronchi P, Allegretti M, Mosalaganti S, et al. (2020) Selective autophagy degrades nuclear pore complexes. Nat Cell Biol 22(2): 159-166.
- Li Z, Mochida K, Nakatogawa H (2024) Macronucleophagy maintains cell viability under nitrogen starvation by modulating micronucleophagy. Nature Communications 15(1): 10670.
- English A, Voeltz G (2013) Endoplasmic reticulum structure and interconnections with other organelles. Cold Spring Harb Perspect Biol 5(4): a013227.
- Liao Y, Duan B, Zhang Y, Zhang X, Xia B, et al. (2019) Excessive ER-phagy mediated by the autophagy receptor FAM134B results in ER stress, the unfolded protein response, and cell death in HeLa cells. J Biol Chem 294(52): 20009-20023.
- Carmona-Gutierrez D, Zimmermann A, Kainz K, Pietrocola F, Chen G, et al. (2019) The flavonoid 4,4'-dimethoxychalcone promotes autophagy-dependent longevity across species. Nat Commun 10(1): 651.
© 2025 Zunxi Huang. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






