- Submissions

Full Text
Integrative Journal of Conference Proceedings
From Environment to Epigenome: Understanding How Social Factors Shape Human Health
Swarup K. Chakrabarti1* and Dhrubajyoti Chattopadhyay1,2
1HP Ghosh Research Center, New Town, Kolkata, West Bengal, India
2Sister Nivedita University, New Town, West Bengal, India
*Corresponding author:Swarup K. Chakrabarti, HP Ghosh Research Center, HIDCO (II), EK Tower, New Town, Kolkata, West Bengal 700161, India
Submission: August 15, 2024;Published: November 19, 2024

Volume4 Issue1November 19, 2024
Abstract
The emerging realm of social epigenomics unveils the intricate interplay between social determinants and chromatin dynamics, offering profound insights into health disparities and disease mechanisms. Social stressors, such as chronic stress and discrimination, serve as catalysts for epigenetic modifications like DNA methylation and histone post-translational modifications (PTMs), which in turn impact stress response pathways and contribute to a spectrum of health conditions. The ramifications of social isolation reverberate on a molecular level, as evidenced by the reshaping of gene networks within critical brain regions linked to emotions and stress responses. Studies also elucidate significant changes in histone acetylation, DNA methylation, and non-coding RNA expression in response to social isolation, shedding light on the complex interplay between social experiences and epigenetic regulation. Beyond social stressors and isolation, factors like nutrition, life style choices, and exposure to environmental toxins wield considerable influence over chromatin dynamics. Positive social interactions and support emerge as crucial mitigating factors against stress-induced epigenetic alterations, not only slowing down epigenetic aging but also emphasizing the importance of fostering supportive social environments for overall well-being. The lasting influence of childhood trauma on the epigenome highlights how early-life experiences can significantly impact mental health throughout one’s lifetime. Adverse childhood events imprint lasting changes on gene expression related to psychiatric disorders and stress management, emphasizing the profound and enduring effects of such experiences. This underscores the critical importance of targeted interventions and policies that address social determinants, aiming to mitigate health disparities and promote overall well-being on a global scale in a comprehensive manner.
Introduction
Exploring environmental epigenetics reveals how a range of external factors-from social adversity to trauma and discrimination-profoundly impact gene expression regulation [1- 3]. These environmental influences intersect with complex societal structures, exacerbating disparities in biological health. Despite advances in epigenetic research, understanding how social determinants of health (SDOH), such as socioeconomic status, education, occupation, and healthcare access, dynamically alter gene expression through epigenetic mechanisms remains underestimated. Recognizing and addressing this complex interplay between environmental epigenetics and social determinants is crucial for mitigating global health disparities [4-6].
Marginalized groups often confront disproportionate health challenges, perpetuating cycles of disadvantage rooted in trauma and discrimination. At the forefront of comprehending these disparities lies the burgeoning field of “social epigenomics” situated at the confluence of genetics, population health, and precision medicine. This field assumes significance as it delves into the social dimensions within epigenomics, furnishing crucial insights into the varied disease manifestations among individuals with identical genetic backgrounds [4-8].
For instance, the variance in health outcomes observed among genetically identical twins raised in disparate socioeconomic contexts underscores the profound influence of social factors on epigenetic regulation and subsequent health outcomes [9,10]. This highlights the pivotal role of epigenetics over genetics in molding health disparities influenced by socioeconomic status (SES). Consequently, social epigenomics elucidates how social, environmental, and genetic factors converge to shape gene expression and disease risk.
Understanding the impact of social and environmental stressors on gene expression is crucial for devising effective interventions and policies to address health disparities. By unraveling these complexities, targeted approaches can be developed that account for the multifaceted interplay between genetics, environment, and social dynamics in shaping health outcomes.
Against this backdrop, the upcoming section seeks to delve into the evidence and underlying mechanisms by which environmental factors, influenced by social dynamics, impact the human epigenome. This understanding could provide a clear avenue for intervening in social influences on human diseases through dietary and nutritional interventions.
Social determinants of epigenetic health: Bridging macro and molecular realms
Social structures wield significant sway over our existence, molding facets such as SES, resource accessibility, and exposure to an array of stressors. Within this intricate tapestry, a multitude of factors intertwine with epigenetic mechanisms, exerting profound influences on our biological constitution. For example, residing in a polluted or tumultuous neighborhood can subject individuals to environmental toxins, auditory disruptions, and social unrest, precipitating epigenetic alterations linked to heightened stress reactivity and heightened disease susceptibility [11,12]. Furthermore, access to healthcare, education, and employment opportunities, all dictated by social structures, modulates epigenetic profiles through differential exposure to stressors and resources [4-8].
Additionally, research findings underscore that the ramifications of childhood poverty extend far beyond immediate financial hardships. Investigations reveal enduring health impacts stemming from poverty, mediated through diverse pathways. Among these pathways, epigenetic alterations emerge as a prominent mechanism whereby environmental conditions, including SES, wield influence over gene expression independently of DNA sequence modifications. Early exposure to poverty correlates with epigenetic modifications in genes governing crucial metabolic processes, such as insulin sensitivity, lipid metabolism, and energy homeostasis [13,14]. These epigenetic changes may predispose individuals to metabolic disorders like Type 2 diabetes (T2D), dyslipidemia, and obesity later in life [15,16].
Moreover, the reverberations of early-life poverty can resonate across generations, potentially perpetuating epigenetic modifications that are transmitted to offspring. This transgenerational transmission of altered gene expression may fuel a cycle of diminished health outcomes and persistent socioeconomic adversity [17]. Furthermore, during adolescence, these epigenetic alterations possess the capacity to mold neural circuitry, modulate emotional reactivity, and shape cognitive functions, thereby setting the stage for future trajectories of both physical and mental health [18,19].
The influence of discrimination on DNA methylation vividly illustrates the profound consequences of societal inequities on our molecular constitution. This nexus underscores the nuanced interconnection between social dynamics and genetic mechanisms, emphasizing how encounters with discrimination can etch enduring signatures on the epigenome, orchestrating alterations in gene expression profiles and exacerbating health disparities [4- 8,20,21].
Examining the influence of social factors on chromatin dynamics
The chromatin, found within the nucleus of eukaryotic cells, represents a dynamic amalgamation of DNA and proteins, forming chromosomes. Central to its structure are nucleosomes, comprised of DNA-enveloping histone proteins. These nucleosomes aggregate into higher-level formations termed chromatin fibers, which exhibit diverse conformations, including the compact 30-nanometer fiber. Euchromatin, characterized by open and accessible regions, facilitates active gene expression, whereas heterochromatin, exhibiting condensed regions, remains transcriptionally silent. Notably, the organization and accessibility of chromatin exert pivotal influences on gene regulation, impacting various cellular processes. Although genetic and environmental factors have traditionally been acknowledged as drivers of chromatin dynamics, recent investigations have unveiled the relevance of social factors in this intricate interplay [22-24].
Maintaining homeostasis necessitates continuous cellular adjustments involving the regulation of gene expression through modulating chromatin dynamics in response to environmental fluctuations [25,26]. In response to environmental shifts, cells must adapt their gene expression patterns accordingly. This adaptation often entails modifying chromatin structure, wherein certain stressors may prompt chromatin relaxation, rendering specific genes more accessible for transcription [27,28]. Conversely, in other instances, chromatin may condense, silencing particular genes. Such cellular adaptive responses play a crucial role in safeguarding human health amid unpredictable environmental changes [29].
Prolonged adaptive responses typically result in predictable alterations in gene expression, stored as memory to finely adjust epigenome regulation in response to recurring stimuli [30,31]. Some of these epigenetic modifications exhibit stability, persisting beyond the initial exposure and forming the basis of epigenetic memory. Through mechanisms like DNA methylation maintenance or propagation of histone post-translational modifications (PTMs) during cell division, cells “remember” past environmental exposures, retaining altered gene expression patterns. This epigenetic memory enhances cells’ ability to respond efficiently to recurring stimuli [32,33].
Importantly, by storing information regarding prior encounters, cells can anticipate future challenges and mount faster and more robust adaptive responses [34]. This predictive capacity is crucial for optimizing cellular function and ensuring survival in fluctuating or challenging environments. Conversely, if cells fail to adapt swiftly to environmental changes, they may struggle to differentiate between the actual environment and their perception of it, influenced by prior epigenetic experiences [35,36].
For instance, the “thrifty phenotype” theory posits that when individuals undergo undernourishment during fetal and early postnatal periods owing to adverse social circumstances such as famine or starvation, their bodies adjust by becoming more epigenetically adaptable [37]. This enhanced adaptability may elevate the susceptibility to chronic illnesses such as metabolic syndrome, obesity, T2D, hypertension, and coronary heart disease in later life. Moreover, these impacts might potentially be inherited by offspring through transgenerational epigenetic inheritance. Even when nutritional constraints are not present, individuals who have experienced social adversity like famine still encounter susceptibility because of their difficulty distinguishing between the real and perceived environment [38].
Telomeres are akin to protective caps found at the chromosome ends, consisting of repetitive DNA sequences and specialized proteins that safeguard our genetic information during cell division [39]. Naturally, telomeres shorten with each cell division, functioning as a molecular clock that reflects cellular aging. Research suggests that social stressors can accelerate this shortening process, potentially by affecting the regulation of telomere-related mechanisms [40]. Moreover, there’s evidence indicating that epigenetic modifications can influence both telomere length and the activity of telomerase, an enzyme responsible for maintaining telomeres [41]. This connection underscores how social factors can influence the epigenetic mechanisms that, in turn, impact telomere maintenance, offering insights into the interplay between our social experiences and molecular biology.
Experiencing the trauma of war and violence doesn’t just impact society and individuals temporarily; it leaves lasting marks that can echo through generations [42]. Investigating these intergenerational effects involves examining how the Holocaust may have altered the epigenetic makeup of survivors’ descendants [43,44]. Through this exploration, the field of social epigenomics can offer profound insights into how historical trauma continues to affect the health and well-being of future generations, shedding light on the underlying mechanisms at play.
Moreover, the COVID-19 pandemic has subjected individuals worldwide to various forms of stress, ranging from fear of infection and financial strain to the disruption of daily routines and social connections [45]. These stressors can activate molecular pathways within cells that modulate gene activity through epigenetic modifications. Thus, the emerging field of social epigenomics holds promise for uncovering the intricate epigenetic changes triggered by the unprecedented stressors of the ongoing COVID-19 pandemic and the extended periods of social isolation it has necessitated.
Hence, in recent years, scientists have been exploring the concept of social epigenomics, inspired by significant studies that have linked social factors to the epigenome. The prevailing perspective depicts social epigenomics as a conceptual framework that suggests the genome, which remains unchanged, is supplemented by a more adaptable epigenome [46]. This epigenome has the capacity to react to various environmental cues, including social factors.
Evidence and mechanisms elucidating how social factors influence chromatin dynamics
The burgeoning field of social epigenomics has unearthed substantial evidence indicating that social factors wield a profound influence on chromatin dynamics, thereby shaping gene expression profiles central to various aspects of health, behavior, and development [4-8]. Despite the predominantly uncharted territory surrounding the relationship between social factors and chromatin interactions, an increasing number of studies are forging connections between adverse social exposures and epigenetic processes, particularly DNA methylation [4-8,47]. This association hints at a potential pathway through which health disparities may manifest.
Nevertheless, our comprehension of epigenetic mechanisms beyond DNA methylation within this framework remains limited, necessitating further exploration. Understanding how social factors sculpt chromatin dynamics holds extensive implications for public health interventions and social policies geared towards fostering health equity. Further research in this area could provide crucial insights that inform strategies to address and reduce health disparities.
Therefore, the following section delves into the evidence
and fundamental mechanisms by which social factors regulate
chromatin dynamics. Here are several pathways through which
social factors can impact chromatin dynamics:
Stress response: Social stressors, like social isolation or
challenges in social hierarchies, can prompt the activation of
stress response pathways in organisms. These pathways often
involve adjustments to chromatin structure through epigenetic
modifications, such as changes in DNA methylation patterns or
histone modifications [48,49].
For instance, research has shown that individuals experiencing chronic stress or trauma frequently exhibit altered DNA methylation patterns, particularly in genes linked to stress response. In a recent study, a locus in the Kit ligand gene (KITLG; cg27512205) displayed the highest correlation with cortisol stress reactivity associated with stress (P=5.8×10-6) in a genome-wide examination of blood DNA methylation in 85 healthy adults [50]. Moreover, evidence supporting the functional relevance of KITLG methylation for regulating stress response in the human brain includes its genomic location within an H3K27ac (histone 3 lysine 27 acetylation) enhancer mark and the correlation between methylation in the blood and the prefrontal cortex [50,51].
Similarly, encounters with discrimination and social disadvantage have been tied to alterations in histone modifications, which can affect gene expression [52]. Intriguingly, the hypothalamus-pituitary- adrenal (HPA) axis, a network of neuroendocrine structures regulating adaptive responses to stress, undergoes changes in gene expression in response to stressors [53]. This includes shifts in the expression of genes encoding corticotrophin-releasing factor (CRF), vasopressin, and adrenocorticotropic hormone (ACTH), disrupting its negative feedback response to stress.
Essentially, stress can induce changes in DNA methylation patterns, influencing the activity of genes involved in behavior regulation. For instance, studies using mouse models of early life stress and chronic social defeat stress have revealed decreased methylation levels within the promoter and regulatory domains of vasopressin and CRF genes in the paraventricular nucleus of the hypothalamus, ultimately resulting in increased expression of these genes [54,55]. Similarly, chronic stress reduces the expression of Bdnf, a gene associated with neuroplasticity and neurogenesis, leading to depression-like behavior in rodents [56]. Although the precise mechanisms behind stress-induced alterations in gene expression remain unclear, one potential molecular mechanism implicated in these changes is DNA methylation.
One proposed mechanism involves the activity of enzymes responsible for adding or removing epigenetic marks. Stress and other environmental factors can directly influence the activity of these enzymes, resulting in alterations in epigenetic patterns. For example, in the nucleus accumbens (Nac), a brain region implicated in motivated and emotional behaviors, certain histone deacetylases (HDACs) such as HDAC2, HDAC3, and HDAC5 have been associated with stress-induced depressive-like behaviors [57,58].
Moreover, external factors, including stress, can indirectly impact epigenetic marks by modulating the availability of cofactors or nutrients essential for the enzymes’ function. For instance, excess cortisol levels during prolonged stress can affect health through their influence on epigenetic processes [59,60]. Cortisol can interact with specific cellular receptors, leading to changes in gene expression. These changes can occur via various mechanisms, including direct interactions with DNA and indirect effects on the enzymes responsible for adding or removing epigenetic marks [61].
These alterations in gene expression can disrupt the production of proteins involved in stress response, immune function, and other essential processes, potentially contributing to the onset of various health conditions.
Moreover, chronic stress can indirectly impact epigenetic processes by influencing behaviors like diet, exercise, and sleep, all of which can affect epigenetic markers. Stress-induced changes in appetite and dietary preferences, for instance, can alter nutrient intake, thus impacting the availability of methyl donors and other compounds crucial for epigenetic processes [62-64].
Similarly, disruptions in sleep patterns, common in individuals experiencing chronic stress, can disturb the expression of genes involved in circadian rhythm regulation and other physiological functions [65]. For example, studies conducted on the pSoBid cohort, a research group in Glasgow with notable social health disparities, have unveiled a correlation between global DNA methylation and SES [66]. Their findings highlighted a connection between severe deprivation, manual labor, and widespread hypomethylation. Intriguingly, they also noted a relationship between global DNA methylation and years of education.
These insights underscore the complex interplay among socioeconomic factors, environmental stressors, and epigenetic mechanisms, emphasizing the multifaceted nature of the link between social determinants and epigenetic processes.
Drosha serves as a crucial RNase-type III protein involved in the intricate process of microRNA biogenesis [67]. It functions by cleaving the lower stem loop of pre-microRNAs, facilitating their transit from the nucleus to the cytoplasm for subsequent maturation into mature microRNAs. Notably, there exists a correlation between the expression of drosha in the hippocampus and depressive behavior in mice [68,69]. Several studies seem to suggest that chronic social defeat stress (SDS) could lead to alterations in DNA methylation within the intron of drosha, potentially resulting in reduced expression levels [70,71]. However, additional replication is necessary to verify this observation.
That being said, the impact of alterations in DNA methylation might be counteracted by additional epigenetic processes. While a robust association between changes in DNA methylation patterns and gene expression has been previously documented, as evidenced by some research, other studies have suggested that certain stress paradigms may be more heavily influenced by alternative epigenetic mechanisms in regulating gene expression. For instance, one investigation revealed an increase in microRNAs targeting Nr3c1, Nr3c2, and Fkbp4 expression following repeated social defeat [72,73]. These findings underscore the potential for other epigenetic mechanisms to modify or supersede the influence of DNA methylation patterns on gene expression level’s.
The hippocampus, a brain region implicated in depression, has been a focus of research attention [74,75]. Numerous studies conducted in mice and rats have shed light on the role of histone acetylation and deacetylation processes within the hippocampus in the development of stress-induced depressive-like behaviors [76- 78].
In experiments involving C57BL/6J male mice subjected to SDS, researchers observed reductions in social interaction and sucrose preference [79,80]. Concurrently, they noted a transient surge followed by a sustained decline in H3K14ac (histone H3 lysine 14 acetylation) levels within the hippocampus. This reduction in sucrose preference suggests a diminished capacity for experiencing pleasure or reward, a common behavioral symptom associated with depressive-like states in rodents [79-81].
Various investigations have highlighted the effectiveness of multiple classes of HDACs in mitigating the effects of SDS. Other functions of histone methylation, an alternative form of PTM for histones, have been implicated in depressive-like behaviors in both rodents and humans. For example, SDS in mice resulted in an increase in H3K27me2 (histone 3 lysine 27 methylation) levels, a transcriptionally repressive epigenetic mark, at specific Bdnf promoter sites (III and IV) within the hippocampus [82]. This increase coincided with a reduction in Bdnf transcript abundance. Notably, this modification persisted over an extended period, remaining at the Bdnf promoter for up to a month following SDS [83].
In the postmortem prefrontal cortex of individuals diagnosed with major depressive disorder (MDD), BDNF mRNA expression exhibited a decrease, while H3K27me3 levels showed an increase at BDNF promoter IV compared to both control subjects and individuals with MDD who had a history of antidepressant use [84,85].
Together, although DNA methylation is commonly viewed as the predominant epigenetic mechanism through which social stressors modulate depressive behaviors, accumulating evidence indicates the involvement of alternative epigenetic pathways, including PTMs and potentially microRNAs, in mediating the stress response. Nevertheless, additional human studies are imperative to corroborate these findings.
Social isolation: The profound influence of social isolation on molecular alterations, encompassing chromatin dynamics, cannot be downplayed, especially amid significant upheavals like pandemics, which frequently precipitate prolonged periods of social seclusion [86]. The repercussions reverberate through mental health and beyond, affecting physiology, cognition, and behavior in profound ways. This phenomenon isn’t confined to humans alone; it spans across species, from Drosophila to mammals [87,88].
For instance, in drosophila, just four days of social isolation trigger significant shifts in the expression of 90 genes, predominantly associated with the immune response [89]. This mirrors broader observations linking social isolation to altered immune function and inflammation, which are strongly implicated in depressive-like behaviors in animal models and clinical depression in humans [90].
Moreover, the effects of social isolation extend beyond gene expression, delving into the intricate world of epigenetics. Studies have illuminated how it can reshape histone acetylation and DNA methylation patterns in the brain, consequently influencing gene expression crucial for social behavior and stress response. This underscores the multifaceted nature of social isolation’s impact, transcending mere social dynamics to fundamentally alter molecular mechanisms underlying mental well-being [91,92].
In fact, in rats, enduring social isolation spanning 6 to 12 weeks triggers significant shifts in gene expression within both the cortex and the nucleus accumbens shell (NAcSh), a critical brain region implicated in processing emotional stimuli [93]. Notably, in the NAcSh, chronic social isolation in adults drives the upregulation of numerous genes, including those encoding transcription factors and epigenetic modifiers like HDAC4 [93]. This implies that prolonged social isolation has the potential to fundamentally reshape gene regulatory networks by modulating the levels of activity-dependent transcription factors and chromatin-modifying proteins.
In rodents, social isolation can also impact the expression of non-coding RNAs, such as miRNAs. Extended isolation during the postnatal period in rats led to distinct changes in miRNA expression within the anterodorsal bed nucleus of the stria terminalis (adBNS), a region associated with anxiety responses [93,94].
There is additional evidence, extending beyond the confines of this review article, illuminating the pivotal connections between social isolation and epigenetic alterations.
Nutrient and lifestyle: The health of an individual, beyond their genetic blueprint, is influenced by a multitude of environmental factors, beginning even before birth and acting through epigenetic mechanisms. Among these factors, nutrition emerges as one of the most extensively studied and understood contributors [95,96]. There is evidence linking inadequate prenatal nutrition to compromised postnatal health and increased susceptibility to diseases. Nutrients exert their influence by either directly inhibiting key epigenetic enzymes like DNMT (DNA methyltransferase), HDAC, or HAT (histone acetyl transferase) or by modulating the availability of essential substrates required for these enzymatic processes.
Social factors, such as the accessibility of nutritious food, the availability of exercise facilities, and exposure to environmental toxins, hold substantial sway over chromatin dynamics. A diet abundant in methyl donors such as folate and methionine can induce notable alterations in DNA methylation patterns, potentially molding gene expression and thereby influencing health outcomes [97,98]. Conversely, contaminants in the environment can disrupt histone modifications, disturbing the intricate equilibrium of chromatin structure and function [11,12].
Thus, our societal milieu plays a pivotal role in sculpting the epigenetic terrain, with repercussions that transcend individual lifestyles and penetrate deeply into the essence of our genetic expression.
Social support: Conversely, the significance of social support and positive interactions on chromatin dynamics cannot be overlooked. Extensive research has revealed a protective influence wielded by nurturing social connections over the orchestration of chromatin regulation. Studies present compelling evidence linking social support to shifts in gene expression pathways crucial for immune function and stress response, with these alterations mediated through epigenetic mechanisms [99,100].
For example, a study revealed that individuals reporting elevated levels of social support demonstrated reduced expression of inflammatory genes, particularly those linked to inflammation and immune activation [101,102]. This suppression was paralleled by alterations in DNA methylation patterns, implying an epigenetic mechanism at play in the impact of social support on the expression of immune-related genes.
Furthermore, a longitudinal study meticulously followed a cohort of individuals over an extensive timeframe, revealing a fascinating correlation: those who reported higher levels of social support displayed a slowdown in epigenetic aging, as indicated by DNA methylation-based biomarkers [103,104]. This suggests that positive social interactions could potentially act as a buffer against the epigenetic changes associated with aging and stress.
In concert, the impact of social support on chromatin dynamics transcends mere psychological well-being, delving deep into the intricate molecular mechanisms governing gene expression and cellular function. These revelations underscore the profound influence of social relationships on human health at a molecular level, underscoring the critical importance of nurturing supportive social environments for overall well-being.
Trauma and adversity: The imprint of trauma and adversity during early life stages resonates deeply within the intricate framework of chromatin dynamics, leaving a lasting mark that echoes through the years. Adverse childhood experiences, ranging from abuse and neglect to household dysfunction, have emerged as potent determinants shaping the epigenetic landscape [3-6].
A groundbreaking study delved into the correlation between childhood maltreatment and DNA methylation patterns in later adulthood. The findings revealed striking evidence of modified DNA methylation in distinct genomic areas among individuals who had endured childhood trauma, especially within genes crucial for stress regulation and mental well-being [105].
Moreover, a comprehensive meta-analysis pooled data from numerous studies exploring the epigenetic repercussions of childhood adversity. The analysis unveiled consistent patterns of DNA methylation alterations linked to early-life trauma across diverse populations and contexts, underscoring the resilience of these epigenetic changes as indicators of adversity [106].
Histone PTMs, pivotal for regulating chromatin dynamics, are profoundly influenced by early-life trauma. A seminal study illuminated the enduring impact of stress experienced in infancy, revealing how it instigates persistent alterations in histone acetylation patterns within the brain [107]. These changes subsequently drive shifts in gene expression patterns associated with psychiatric disorders such as depression and anxiety.
Figure 1:(A) Environmental changes, such as global warming and its associated effects, directly contribute to social adversity. (B) Environment-induced alterations exert a direct influence on the human epigenome, primarily through the regulation of chromatin structure and function. The chromatin, which encompasses DNA and its associated proteins, serves as a crucial mediator of gene expression. These epigenetic changes play a pivotal role in shaping an individual’s response to environmental stimuli and can have implications for health outcomes. (C) Conversely, social adversity stemming from various social factors also exerts an impact on the epigenome. Socioeconomic disparities, discrimination, and lack of access to healthcare are among the determinants of social adversity that contribute to epigenetic modifications. Collectively, this interconnectedness underscores the complex interplay between environmental, social, and biological factors in shaping the human epigenome and ultimately influencing health and disease outcomes. The final figure is created by combining individual images obtained from web scraping on Google.
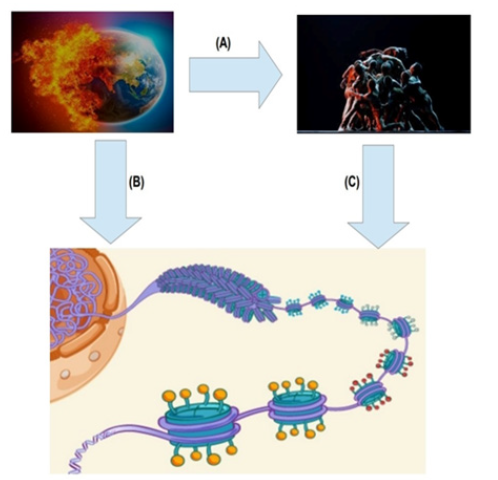
Together, these discoveries emphasize the significant influence of early-life trauma and adversity on chromatin dynamics, offering a molecular framework for understanding their lasting effects on mental health and stress response. By unraveling the epigenetic mechanisms driving these effects, efforts are made not only to illuminate the intricate interplay between the environment and gene regulation but also to pave the way for targeted interventions to alleviate the enduring consequences of childhood adversity (Figure 1).
Future Direction
Future endeavors should incorporate the utilization of systems biology, offering a comprehensive framework to grasp the intricate interplay among environmental changes, social dynamics, and the health of epigenetics. By merging omics data with sophisticated computational modeling, the intricate molecular conversations within biological systems can be unraveled [108]. This enables the anticipation of system responses to stressors or illnesses, leveraging techniques like network analysis and machine learning. Such models simulate environmental influences, predict epigenetic alterations over generations, and assess intervention strategies.
Computational models are vital for understanding how epigenetic mechanisms respond to environmental cues like DNA methylation and histone PTMs. By simulating these responses, insight is gained into how molecular changes affect gene expression patterns and individual health [109]. This informs targeted interventions to mitigate health issues stemming from these molecular roots.
Longitudinal studies spanning generations and diverse environments are crucial for understanding how environmental and social factors affect epigenetic health over time. Tracking individuals over time can reveal how lifestyle changes impact epigenetic mechanisms and health outcomes, informing personalized healthcare. Comparing groups facing different social conditions and interventions can show how alleviation measures affect epigenetic markers, shedding light on the effectiveness of social interventions like education and healthcare in reversing or mitigating epigenetic changes caused by adversity.
Reliable epigenetic biomarkers linked to environmental alterations and social adversity are crucial for understanding related diseases over the over the long term [110]. These biomarkers are stable and reproducible modifications or mechanisms detectable in body fluids or tissues. They serve various purposes, including disease detection, risk prediction, and therapy monitoring. Epigenetic biomarkers like microRNAs and histone PTMs persist in bodily fluids, highlighting their importance in quantifying health impacts from environmental changes.
Engaging communities directly in research efforts fosters trust, promotes inclusivity, and ensures that interventions are tailored to local needs and realities. Community-based participatory research approaches empower marginalized populations to actively contribute to the co-creation of knowledge and the development of solutions, thereby enhancing the relevance and effectiveness of interventions.
Integrating findings from molecular research into policy frameworks is crucial for translating scientific knowledge into actionable policies that address the root causes of health disparities exacerbated by environmental changes. Advocating for evidencebased policies that prioritize environmental justice, health equity, and sustainable development can catalyze meaningful change at local, national, and global levels.
Increasing public awareness of the connections between environmental changes, social factors, and epigenetic health is crucial for informed decision-making. Through science communication, education, and community outreach, individuals can make choices that benefit both their personal health and the environment. This fosters resilience and sustainability, enhancing understanding of the interplay between environmental shifts, social dynamics, and genetic expression.
Last but not least, by employing epitherapeutics to stabilize altered epigenomes, we have the potential to significantly impact molecular medicine and population health on a broad scale.
Funding
Funding for the research project comes from an intramural grant provided by the Bandhan Group. This grant is specifically intended to support the activities of the H. P. Ghosh Research Center located in Kolkata, India.
Conflict of Interest
The authors declare that they have no conflicts of interest to disclose regarding the publication of this research.
References
- Breton CV, Landon R, Kahn LG, Enlow MB, Peterson AK, et al. (2021) Exploring the evidence for epigenetic regulation of environmental influences on child health across generations. Commun Biol 4(1): 769.
- Tung J, Gilad Y (2013) Social environmental effects on gene regulation. Cell Mol Life Sci 70(22): 4323-4339.
- Non AL (2021) Social epigenomics: Are we at an impasse? Epigenomics 13(21): 1747-1759.
- Chelak K, Chakole S (2023) The role of social determinants of health in promoting health equality: A narrative review. Cureus 15(1): e33425.
- Islam MM (2019) Social determinants of health and related inequalities: Confusion and implications. Front Public Health 7: 11.
- Sulley S, Bayssie M (2021) Social determinants of health: An evaluation of risk factors associated with inpatient presentations in the United States. Cureus 13(2): e13287.
- Martin CL, Ghastine L, Lodge EK, Dhingra R, Caviness CKW (2022) Understanding health inequalities through the lens of social epigenetics. Annu Rev Public Health 43: 235-254.
- Mancilla VJ, Peeri NC, Silzer T, Basha R, Felini M, et al. (2020) Understanding the interplay between health disparities and epigenomics. Front Genet 11: 903.
- Wong CC, Caspi A, Williams B, Craig IW, Houts R, et al. (2010) A longitudinal study of epigenetic variation in twins. Epigenetics 5(6): 516-526.
- Krieger N, Chen JT, Coull BA, Selby JV (2005) Lifetime socioeconomic position and twins' health: An analysis of 308 pairs of United States women twins. PLoS Med 2(7): e162.
- Mukherjee S, Dasgupta S, Mishra PK, Chaudhury K. (2021) Air pollution-induced epigenetic changes: Disease development and a possible link with hypersensitivity pneumonitis. Environ Sci Pollut Res Int 28(40): 55981-56002.
- Grova N, Schroeder H, Olivier JL, Turner JD (2019) Epigenetic and neurological impairments associated with early life exposure to persistent organic pollutants. Int J Genomics 2019: 2085496.
- Scorza P, Duarte CS, Hipwell AE, Posner J, Ortin A, et al. (2019) Research review: Intergenerational transmission of disadvantage: Epigenetics and parents' childhoods as the first exposure. J Child Psychol Psychiatry 60(2): 119-132.
- Arnold NS, Hooten NN, Zhang Y, Lehrmann E, Wood W, et al. (2020) The association between poverty and gene expression within peripheral blood mononuclear cells in a diverse Baltimore City cohort. PLoS One 15(9): e0239654.
- Loh M, Zhou L, Ng HK, Chambers JC (2019) Epigenetic disturbances in obesity and diabetes: Epidemiological and functional insights. Mol Metab 27S(Suppl): S33-S41.
- Long Y, Mao C, Liu S, Tao Y, Xiao D (2024) Epigenetic modifications in obesity-associated diseases. Med Comm (2020) 5(2): e496.
- Blakemore SJ, Den Ouden H, Choudhury S, Frith C (2007) Adolescent development of the neural circuitry for thinking about intentions. Soc Cogn Affect Neurosci 2(2): 130-139.
- Jagtap A, Jagtap B, Jagtap R, Lamture Y, Gomase K (2023) Effects of prenatal stress on behavior, cognition, and psychopathology: A comprehensive review. Cureus 15(10): e47044.
- Mendoza VBD, Huang Y, Crusto CA, Sun YV, Taylor JY (2018) Perceived racial discrimination and DNA methylation among African American women in the interGEN study. Biol Res Nurs 20(2): 145-152.
- Santos HP, Nephew BC, Bhattacharya A, Tan X, Smith L, et al. (2018) Discrimination exposure and DNA methylation of stress-related genes in Latina mothers. Psychoneuroendocrinology 98: 131-138.
- Maeshima K, Imai R, Tamura S, Nozaki T (2014) Chromatin as dynamic 10-nm fibers. Chromosoma 123(3): 225-237.
- Alberts B, Johnson A, Lewis J (2002) Molecular biology of the cell. (4th edn), Garland science; Chromosomal DNA and its packaging in the chromatin fiber.
- Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403(6765): 41-45.
- Ordog T, Syed SA, Hayashi Y, Asuzu DT (2012) Epigenetics and chromatin dynamics: A review and a paradigm for functional disorders. Neurogastroenterol Motil 24(12): 1054-1068.
- Ahuir AP, Torró JF, Proft M (2020) Capturing and understanding the dynamics and heterogeneity of gene expression in the Living cell. Int J Mol Sci 21(21): 8278.
- Fang L, Wuptra K, Chen D, Li H, Huang SK, et al. (2014) Environmental-stress-induced chromatin regulation and its heritability. J Carcinog Mutagen 5(1): 22058.
- Ito S, Das ND, Umehara T, Koseki H (2022) Factors and mechanisms that influence chromatin-mediated enhancer-promoter interactions and transcriptional regulation. Cancers (Basel) 14(21): 5404.
- Smale ST, Fisher AG (2002) Chromatin structure and gene regulation in the immune system. Annu Rev Immunol 20: 427-462.
- Shang A, Bieszczad KM (2022) Epigenetic mechanisms regulate cue memory underlying discriminative behavior. Neurosci Biobehav Rev 141: 104811.
- Mehler MF (2008) Epigenetic principles and mechanisms underlying nervous system functions in health and disease. Prog Neurobiol 86(4): 305-341.
- Bruno S, Williams RJ, Vecchio DD (2022) Epigenetic cell memory: The gene's inner chromatin modification circuit. PLoS Comput Biol 18(4): e1009961.
- Saxton DS, Rine J (2019) Epigenetic memory independent of symmetric histone inheritance. Elife 8: e51421.
- Bhargavi G, Subbian S (2024) The causes and consequences of trained immunity in myeloid cells. Front Immunol 15: 1365127.
- Luo X, Song R, Moreno DF, Ryu HY, Hochstrasser M, et al. (2020) Epigenetic mechanisms contribute to evolutionary adaptation of gene network activity under environmental selection. Cell Rep 33(4): 108306.
- Brooks AN, Turkarslan S, Beer KD, Lo FY, Baliga NS (2011) Adaptation of cells to new environments. Wiley Interdiscip Rev Syst Biol Med 3(5): 544-561.
- Hales CN, Barker DJ (2001) The thrifty phenotype hypothesis. Br Med Bull 60: 5-20.
- Feinberg AP, Fallin MD (2015) epigenetics at the crossroads of genes and the environment. JAMA 314(11): 1129-1130.
- Laberthonnière C, Magdinier F, Robin JD (2019) Bring it to an end: Does Telomeres Size Matter? Cells 8(1):30.
- Lin J, Epel E (2022) Stress and telomere shortening: Insights from cellular mechanisms. Ageing Res Rev 73: 101507.
- Dogan F, Forsyth NR (2021) Epigenetic features in regulation of telomeres and telomerase in stem cells. Emerg Top Life Sci 5(4):497-505.
- Almoshmosh N (2016) The role of war trauma survivors in managing their own mental conditions, Syria civil war as an example. Avicenna J Med 6(2):54-59.
- Dashorst P, Mooren TM, Kleber RJ, De Jong PJ, Huntjens RJC (2019) Intergenerational consequences of the Holocaust on offspring mental health: A systematic review of associated factors and mechanisms. Eur J Psychotraumatol 10(1): 1654065.
- Švorcová J (2023) Transgenerational epigenetic inheritance of traumatic experience in mammals. Genes (Basel) 14(1): 120.
- Koçak O, Koçak ÖE, Younis MZ (2021) The psychological consequences of COVID-19 fear and the moderator effects of individuals' underlying illness and witnessing infected friends and family. Int J Environ Res Public Health 18(4): 1836.
- Deichmann U (2020) The social construction of the social epigenome and the larger biological context. Epigenetics Chromatin. 13(1): 37.
- Parodi L, Mayerhofer E, Narasimhalu K, Yechoor N, Comeau ME, et al. (2023) Social determinants of health and cerebral small vessel disease: Is epigenetics a key mediator? J Am Heart Assoc 12(13): e029862.
- Gudsnuk K, Champagne FA (2012) Epigenetic influence of stress and the social environment. ILAR J 53(3-4): 279-288.
- Soga T, Teo CH, Parhar I (2021) Genetic and epigenetic consequence of early-life social stress on depression: Role of serotonin-associated genes. Front Genet 11: 601868.
- Houtepen LC, Vinkers CH, Carrillo RT, Hiemstra M, Van LPA, et al. (2016) Genome-wide DNA methylation levels and altered cortisol stress reactivity following childhood trauma in humans. Nat Commun 7: 10967.
- Shahhosseini A, Bourova FE, Derakhshan S, Aminishakib P, Goudarzi A (2023) High levels of histone H3 K27 acetylation and tri-methylation are associated with shorter survival in oral squamous cell carcinoma patients. Biomedicine (Taipei) 13(1): 22-38.
- Toraño EG, García MG, Fernández MJL, Niño GP, Fernández AF (2016) The impact of external factors on the epigenome: In utero and over lifetime. Biomed Res Int 2016: 2568635.
- Goncharova ND (2013) Stress responsiveness of the hypothalamic-pituitary-adrenal axis: Age-related features of the vasopressinergic regulation. Front Endocrinol (Lausanne) 4: 26.
- Dedic N, Chen A, Deussing JM (2018) The CRF family of neuropeptides and their receptors - mediators of the central stress response. Curr Mol Pharmacol 11(1): 4-31.
- Persaud NS, Cates HM (2023) The epigenetics of anxiety pathophysiology: A DNA methylation and histone modification focused review. eNeuro 10(4): ENEURO.0109-21.2021.
- Yang T, Nie Z, Shu H, Kuang Y, Chen X, et al. (2020) The role of BDNF on neural plasticity in depression. Front Cell Neurosci 14: 82.
- Qian W, Yu C, Wang S, Niu A, Shi G, et al. (2021) Depressive-like behaviors induced by chronic social defeat stress are associated with HDAC7 reduction in the nucleus accumbens. Front Psychiatry 11: 586904.
- Covington HE, Maze I, LaPlant QC, Vialou VF, Ohnishi YN, et al. (2009) Antidepressant actions of histone deacetylase inhibitors. J Neurosci 29(37): 11451-11460.
- Hunter RG (2012) Epigenetic effects of stress and corticosteroids in the brain. Front Cell Neurosci 6: 18.
- Zannas AS (2019) Epigenetics as a key link between psychosocial stress and aging: Concepts, evidence, mechanisms. Dialogues Clin Neurosci 21(4): 389-396.
- Mourtzi N, Sertedaki A, Charmandari E (2021) Glucocorticoid signaling and epigenetic alterations in stress-related disorders. Int J Mol Sci 22(11): 5964.
- Plaza DJ, Izquierdo D, Torres MÁ, Baig AT, Aguilera CM, et al. (2022) Impact of physical activity and exercise on the epigenome in skeletal muscle and effects on systemic metabolism. Biomedicines 10(1): 126.
- Fernandes J, Arida RM, Gomez PF (2017) Physical exercise as an epigenetic modulator of brain plasticity and cognition. Neurosci Biobehav Rev 80: 443-456.
- Imam MU, Ismail M (2017) The impact of traditional food and lifestyle behavior on epigenetic burden of chronic disease. Glob Chall 1(8): 1700043.
- Phan TX, Malkani RG (2018) Sleep and circadian rhythm disruption and stress intersect in Alzheimer's disease. Neurobiol Stress 10: 100133.
- McGuinness D, McGlynn LM, Johnson PC, MacIntyre A, Batty GD, et al. (2012) Socio-economic status is associated with epigenetic differences in the pSoBid cohort. Int J Epidemiol 41(1): 151-160.
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, et al. (2004) The drosha-DGCR8 complex in primary microRNA processing. Genes Dev 18(24): 3016-3027.
- Mulligan MK, Dubose C, Yue J, Miles MF, Lu L, et al. (2013) Expression, covariation, and genetic regulation of miRNA Biogenesis genes in brain supports their role in addiction, psychiatric disorders, and disease. Front Genet 4: 126.
- Dwivedi Y (2014) Emerging role of microRNAs in major depressive disorder: diagnosis and therapeutic implications. Dialogues Clin Neurosci 16(1): 43-61.
- Hing B, Braun P, Cordner ZA, Ewald ER, Moody L, et al. (2018) Chronic social stress induces DNA methylation changes at an evolutionary conserved intergenic region in chromosome X. Epigenetics 13(6): 627-641.
- Oh YE, Nguyen TB, Rami FZ, Karamikheirabad M, Chung YC (2022) Impact of social defeat stress on DNA methylation in Drd2, Nr3c1, and Stmn1 in wild-type and stmn1 knock-out mice. Clin Psychopharmacol Neurosci 20(1): 51-60.
- Jung SH, Wang Y, Kim T, Tarr A, Reader B, et al. (2015) Molecular mechanisms of repeated social defeat-induced glucocorticoid resistance: Role of microRNA. Brain Behav Immun 44: 195-206.
- Clayton SA, Jones SW, Kurowska SM, Clark AR (2018) The role of microRNAs in glucocorticoid action. J Biol Chem 293(6): 1865-1874.
- Malykhin NV, Carter R, Seres P, Coupland NJ (2010) Structural changes in the hippocampus in major depressive disorder: Contributions of disease and treatment. J Psychiatry Neurosci 35(5): 337-343.
- Tartt AN, Mariani MB, Hen R, Mann JJ, Boldrini M (2022) Dysregulation of adult hippocampal neuroplasticity in major depression: Pathogenesis and therapeutic implications. Mol Psychiatry 27(6): 2689-2699.
- Bonomi RE, Girgenti M, Krystal JH, Cosgrove KP (2022) A role for histone deacetylases in the biology and treatment of post-traumatic stress disorder: What do we know and where do we go from here? Complex Psychiatry 8(1-2): 13-27.
- Peixoto L, Abel T (2013) The role of histone acetylation in memory formation and cognitive impairments. Neuropsychopharmacology 38(1): 62-76.
- Saw G, Tang FR (2020) Epigenetic regulation of the hippocampus, with special reference to radiation exposure. Int J Mol Sci 21(24): 9514.
- Iñiguez SD, Riggs LM, Nieto SJ, Dayrit G, Zamora NN, et al. (2014) Social defeat stress induces a depression-like phenotype in adolescent male c57BL/6 mice. Stress 17(3): 247-255.
- Sial OK, Gnecco T, Cardona AAM, Vieregg E, Cardoso EA, et al. (2021) Exposure to vicarious social defeat stress and western-style diets during adolescence leads to physiological dysregulation, decreases in reward sensitivity, and reduced antidepressant efficacy in adulthood. Front Neurosci 15: 701919.
- Sun H, Kennedy PJ, Nestler EJ (2013) Epigenetics of the depressed brain: role of histone acetylation and methylation. Neuropsychopharmacology 38(1): 124-37.
- Peña CJ, Nestler EJ (2018) Progress in epigenetics of depression. Prog Mol Biol Transl Sci 157: 41-66.
- Castino MR, Baker AD, Ratnu VS, Shevchenko G, Morris KV, et al. (2018) Persistent histone modifications at the BDNF and Cdk-5 promoters following extinction of nicotine-seeking in rats. Genes Brain Behav 17(2): 98-106.
- Yu H, Chen ZY (2011) The role of BDNF in depression on the basis of its location in the neural circuitry. Acta Pharmacol Sin 32(1): 3-11.
- Šalamon AI, Kouter K, Videtič PA (2022) Depressive disorder and antidepressants from an epigenetic point of view. World J Psychiatry 12(9): 1150-1168.
- Arzate MRG, Lottenbach Z, Schindler V, Jawaid A, Mansuy IM (2020) Long-term impact of social isolation and molecular underpinnings. Front Genet 11: 589621.
- Agrawal P, Chung P, Heberlein U, Kent C (2019) Enabling cell-type-specific behavioral epigenetics in Drosophila: A modified high-yield INTACT method reveals the impact of social environment on the epigenetic landscape in dopaminergic neurons. BMC Biol 17(1): 30.
- Chen M, Sokolowski MB (2022) How social experience and environment impacts behavioural plasticity in Drosophila. Fly (Austin) 16(1): 68-84.
- Leschak CJ, Eisenberger NI (2019) Two distinct immune pathways linking social relationships with health: Inflammatory and antiviral processes. Psychosom Med 81(8): 711-719.
- Li P, Yan Z (2023) An epigenetic mechanism of social isolation stress in adolescent female mice. Neurobiol Stress 29: 100601.
- Popa N, Boyer F, Jaouen F, Belzeaux R, Gascon E (2020) Social isolation and enrichment induce unique miRNA signatures in the prefrontal cortex and behavioral changes in mice. iScience 23(12): 101790.
- Siddeek B, Simeoni U (2022) Epigenetics provides a bridge between early nutrition and long-term health and a target for disease prevention. Acta Paediatr 111(5): 927-934.
- Carlberg C, Velleuer E (2023) Nutrition and epigenetic programming. Curr Opin Clin Nutr Metab Care 26(3): 259-265.
- Mahmoud AM, Ali MM (2019) Methyl donor micronutrients that modify DNA methylation and cancer outcome. Nutrients 11(3): 608.
- Crider KS, Yang TP, Berry RJ, Bailey LB (2012) Folate and DNA methylation: A review of molecular mechanisms and the evidence for folate's role. Adv Nutr 3(1): 21-38.
- Cole SW (2009) Social regulation of human gene expression. Curr Dir Psychol Sci 18(3): 132-137.
- Viana BJ, Souza de FB, Antoniazzi V, De Souza DSC, Vedovelli K, et al. (2019) Social isolation and social support at adulthood affect epigenetic mechanisms, brain-derived neurotrophic factor levels and behavior of chronically stressed rats. Behav Brain Res 366: 36-44.
- Trachtenberg E (2024) The beneficial effects of social support and prosocial behavior on immunity and health: A psychoneuroimmunology perspective. Brain Behav Immun Health 37: 100758.
- Murray DR, Haselton MG, Fales M, Cole SW (2019) Subjective social status and inflammatory gene expression. Health Psychol 38(2): 182-186.
- Mareckova K, Pacinkova A, Marecek R, Sebejova L, Izakovicova HL, et al. (2023) Longitudinal study of epigenetic aging and its relationship with brain aging and cognitive skills in young adulthood. Front Aging Neurosci 15: 1215957.
- Grant CD, Jafari N, Hou L, Li Y, Stewart JD, et al. (2017) A longitudinal study of DNA methylation as a potential mediator of age-related diabetes risk. Geroscience 39(5-6): 475-489.
- Mehta D, Klengel T, Conneely KN, Smith AK, Altmann A, et al. (2013) Childhood maltreatment is associated with distinct genomic and epigenetic profiles in posttraumatic stress disorder. Proc Natl Acad Sci USA 110(20): 8302-8307.
- Neves I, Dinis ORJ, Magalhães T (2019) Epigenomic mediation after adverse childhood experiences: A systematic review and meta-analysis. Forensic Sci Res 6(2): 103-114.
- McEwen BS, Nasca C, Gray JD (2016) Stress effects on neuronal structure: Hippocampus, amygdala, and prefrontal cortex. Neuropsychopharmacology 41(1): 3-23.
- Dahal S, Yurkovich JT, Xu H, Palsson BO, Yang L (2020) Synthesizing systems biology knowledge from omics using genome-scale models. Proteomics 20(17-18): e1900282.
- Raghavan K, Ruskin HJ, Perrin D, Goasmat F, Burns J (2010) Computational micromodel for epigenetic mechanisms. PLoS One 5(11): e14031.
- Park HL (2020) Epigenetic biomarkers for environmental exposures and personalized breast cancer prevention. Int J Environ Res Public Health 17(4): 1181.
- García GJL, Seco CM, Tollefsbol TO, Romá MC, Peiró CL, et al. (2017) Epigenetic biomarkers: Current strategies and future challenges for their use in the clinical laboratory. Crit Rev Clin Lab Sci 54(7-8): 529-550.
- Ueda J, Yamazaki T, Funakoshi H (2023) Toward the development of epigenome editing-based therapeutics: Potentials and challenges. Int J Mol Sci 24(5): 4778.
- Khan A, Khan A, Khan MA, Malik Z, Massey S, et al. (2024) Phytocompounds targeting epigenetic modulations: An assessment in cancer. Front Pharmacol 14: 1273993.
- Ahuja N, Sharma AR, Baylin SB (2016) Epigenetic therapeutics: A new weapon in the war against cancer. Annu Rev Med 67: 73-89.
© 2024 Swarup K. Chakrabarti. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






