- Submissions
Full Text
Examines in Marine Biology and Oceanography
Bacterial Community Profiling of the Arabian Sea Oxygen Minimum Zone Sediments using Cultivation Independent Approach
Baby Divya1*, Annie Feby2 and Shanta Nair3
1,2Department of Zoology, St. Xavier's College for Women, India
3Microbiology Laboratory, National Institute of Oceanography, Goa, India
*Corresponding author: Baby Divya, Department of Zoology, St Xavier's College for Women, Kerala, India
Submission: November 02, 2017; Published: December 07, 2017

ISSN: 2578-031X Volume1 Issue1
Abstract
The eastern Arabian Sea has a unique and permanent oxygen minimum zone (OMZ) that extends along the western continental margin of India. In order to understand the bacterial community structure and diversity of OMZ sediment of the Arabian Sea (AS), PCR-DGGE analysis were carried out for samples collected off Ratnagiri, Goa and Karwar at 50m, 200m, 500m and 1000m depths. PCR-DGGE analysis showed that Proteobacteria dominated all the sites ie 50m (non-OMZ) and 200m, 500m and 1000m (OMZ) along the depth gradient. Majority of the bands were attributed to the Proteobacteria. The other groups obtained belonged to Nitrospira, Bacteriodetes, Actinobacteria, Chloroflexi and Acidobacteria. The species diversity index was higher in the OMZ sites compared to the non-OMZ sites.
Keywords: Arabian sea; OMZ; Bacterial diversity; DGGE
Introduction
Oxygen minimum zones are common and permanent features in the most productive regions of all oceans where dissolved oxygen concentrations are as low as 0.5mlL-1, present at different water depths ranging from shelf to upper bathyal zones (101300m). Oxygen depletion has become widespread in the world oceans occurring as permanent, seasonal and episodic features [1]. In the present scenario of the dramatic global climate change, the likelihood of the expansion of OMZs, thereby acidifying the oceans may not be unanticipated, thus posing destructive effects to oceanic ecosystems [2,3]. Of the total OMZ area 59% occurs in Indian Ocean i.e. Arabian Sea and Bay of Bengal [4]. They are associated with the continental margins where burial of organic carbon takes place in the sediments and contribute to the global carbon cycle [5]. The oxygen depleted bottom water affects the characteristics of the sediment pore water as they are in close association [6]. Thus the region where oxygen minimum zone impinges on the sea floor creates a strong gradient in dissolved oxygen concentrations and organic carbon thus serving as a peculiar habitat for the organisms. These gradients are known to strongly influence the composition and structure of prokaryotic assemblages [7]. Culture-dependent methods are known to be inadequate for the analysis of microbial communities because only a small fraction of bacteria in the environmental samples are culturable [8]. The advent of molecular methods based on 16S rRNA genes has now been widely used for providing new insights into microbial diversity and into structure and dynamics of microbial communities. PCR-DGGE of 16S rRNA gene fragments has been found suitable to analyze multiple samples simultaneously and to assess temporal and spatial dynamics of microbial community [9]. In the present study an attempt has been made to understand the spatial variation in bacterial diversity associated with sediments along a depth gradient in the OMZ in three latitudes such as off Ratnagiri, off Goa and off Karwar. This may provide important insights into the composition and variability of bacterial communities in these regions and processes occurring therein.
Methods
Collection of sediments
Sediment samples were collected from the Arabian Sea (AS) off Ratnagiri, Goa, and Karwar at 50m, 200m, 500m and 1000m depths, onboard FORV Sagar Sampada #254. The stations at the latter three sites (200m, 500m and 1000m) fall in the OMZ while the one at 50m depth was a control site which was a coastal station. The samples were handled aseptically and preserved in liquid nitrogen until DNA extraction.
DNA extraction from sediments
DNA was extracted using modification of earlier method [10]. The purified DNA was estimated using Nanodrop spectrophotometer (NanoDrop Technologies, Inc, USA).
PCR-DGGE analysis
Table 1: Coordinates of the sampling stations.
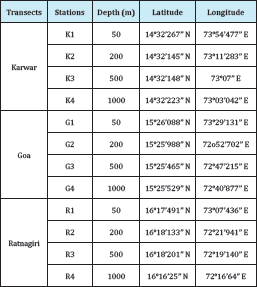
16S rRNA gene amplification of the purified DNA from all the sites was performed using the primers, 341F-CCT ACG GGA GGC AGC AG (59bp GC clamp attached) and 907R-ATT ACC GCG GCT GCT GG respectively [9]. The reaction was carried out using the touchdown protocol-5min at 94 0C, 15 cycles with 1 cycle consisting of 40sec at 94 0C, decreasing of the annealing temperature from 65-55 0C for 40 sec and extension of 72 0C for 40sec. In addition another 20 cycles (1 cycle consisting of 40sec at 940C, 40sec at 50 0C and 1min 30sec at 72 0C ) and a final extension at 72 0C for 5min. The PCR amplification was performed in a Thermal cycler (Eppendorf, Germany). The amplicons were inspected on 1.5% (w/v) agarose gel and visualized using the gel documentation system (Eastman Kodak Company, USA). Sequencing was carried out by a commercial company (Chromous Biotech Pvt. Ltd., India). 16S rRNA gene amplicons were analyzed by DGGE using Dcode Universal Mutation Detection System (Biorad, USA). The samples were mixed with equal amounts of loading dye and loaded on to a 10% acrylamide gel (acrylamide: bisacryamide 37.5:1) in 1X TAE buffer with a 20%- 80% denaturing gradient (100% denaturing gradient containing 7M urea and 40% formamide). Electrophoresis was performed at a constant temperature of 60 0C and a voltage of 100V for 17 hours. The gel was stained with 1X SYBR Gold stain (Molecular Probes, USA) for 15 minutes and photographed with a gel documentation system (Eastman Kodak Company, USA) (Table 1).
Sequencing of DGGE bands and phylogenetic analysis
DGGE bands were excised with a sterile surgical blade, transferred to 10ul of deionized water and incubated at 4 0C for 12 hours. 0.5ul of the supernatant was used as a template after a spin for re-amplification using the primers 341F-CCT ACG GGA GGC AGC AG (GC clamp attached) and 907R-ATT ACC GCG GCT GCT GG respectively [9]. The PCR products were run on 1.5% agarose gel. Further, they were electrophoresed on a parallel gel along with the environmental sample in order to make sure of its position and distinction. The eluted products were also amplified using PCR primer without GC clamp. The resulting PCR products were run on 1.5% agarose gel for assessment of quality and quantity. The PCR products were cloned using TOPO TA cloning kit (Invitrogen, USA). Five positive clones were selected at random and plasmids were extracted using Qiaquick plasmid extraction kit (Qiagen, USA). The 16S rRNA genes in the plasmids were amplified using the vector primers M13F-GTA AAA CGA CGG CCA G and M13R-CAG GAA ACA GCT ATG AC. The PCR products were then sequenced (Chromous Biotech private limited, Bangalore). The sequences obtained were edited using DNA baser (DNA Baser, USA) and compared with sequences in GenBank using Basic Local Alignment Search Tool (BLAST). Phylogenetic tree was constructed using 22 sequences obtained from DGGE analysis based on neighbour-joining method using MEGA software [11].
Nucleotide sequence accession numbers
The partial 16S rRNA gene environmental sequences obtained from this study were deposited in the Genbank database under accession numbers GU968618-GU968638.
Results
DGGE analysis of bacterial community structure
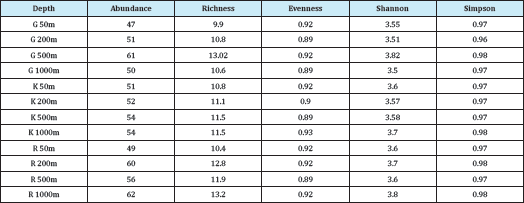
The DGGE profiles obtained for the 12 sampling sites in the three transects i.e. Goa, Karwar and Ratnagiri is shown in Figure 1. The total number of band positions detected was 82 and the number of bands per sample ranged from 47-62 in the all the samples. However, the maximum number of bands was obtained in the OMZ sites 200m, 500m and 1000m at all latitudes. In control sites, the number of bands was more or less similar with 47, 51 and 49 in Goa, Karwar and Ratnagiri respectively. Statistical analysis of the DGGE data showed that at the level of 69% similarity two clusters were formed, which did not show any clustering of similar sites.
The diversity index was also calculated and all the indices showed slight variation among transect and stations it was high in the OMZ sites compared to the non-OMZ sites in all latitudes (Table 2). The Shannon-Weiner diversity index (H') was maximum recorded in Goa 500m with a value of about 3.82. In the Goa transect the Shannon-Weiner index (H') ranged from 3.5 to 3.8, Karwar from 3.5 to 3.7, Ratnagiri 3.6 to 3.8 (Figure 2).
Figure 1: Hierarchical cluster analysis of the 16S rRNA DGGE band profiles of three transects (G-Goa, R-Ratnagiri and K-Karwar) and depths.
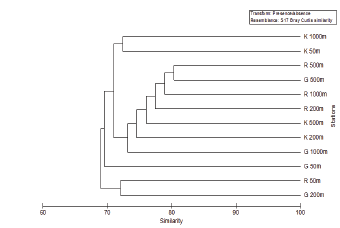
Figure 2: Variation in Shannon-Weiner diversity index at various depths (m).
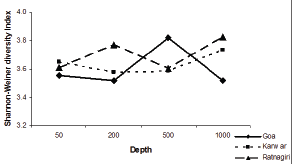
Table 2: Diversity indices of three transect Goa (G), Karwar (K) and Ratnagiri (R).
Phylogenetic analysis of excised bands
Figure 3: Neighbor-joining tree showing the phylogenetic affiliations of bacteria derived from the sequences of excised DGGE bands. The sequences obtained in this study are shown in bold. The bar represents 5% sequence divergence. Planctomyces maris was used as the out group.
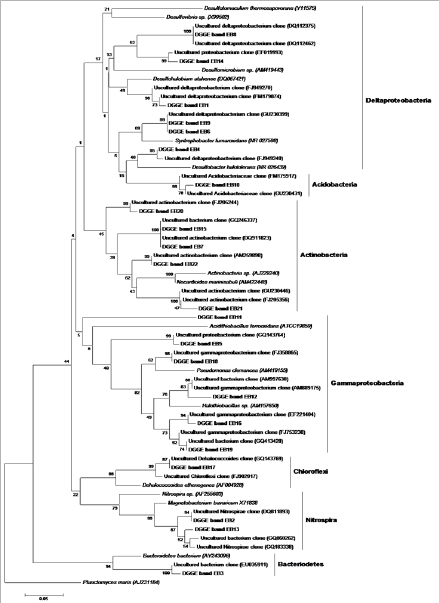
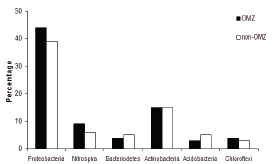
Twenty two bands were excised from gels corresponding to the different stations in the three transect based on the relative band intensity. Same results were obtained for the bands that were excised from the same position, but in different lanes. Unique bands detected at different sites were also excised. Only dominant bands were sequenced that produced good results, suggesting the presence of other bacteria that may be present in lower numbers. The phylogenetic analysis revealed that the sequences belonged to Proteobacteria, Acidobacteria, Actinobacteria, Chloroflexi, Nitrospira and Bacteriodetes (Figure 3). The number of phylogenetic groups (> 4) was higher in the OMZ region. Proteobacteria was the dominant phylum at all depths with deltaproteobacteria and gammaproteo bacteria forming the major classes. All the groups were invariably observed at all the depths without any differentiation of the OMZ and non-OMZ regions. In the OMZ region the density of the phyla such as Proteobacteria, Nitrospira and Chloroflexi were higher than the non-OMZ region (Figure 4). Proteobacteria accounted for 44% of the population in the OMZ as compared to 39% in the non-OMZ. Nitrospira and Chloroflexi comprised of 9% and 4% respectively in the OMZ and 6% and 3% respectively in the non-OMZ region. However, in the non-OMZ region Bacteriodetes and Acidobacteria, each contributing to 5% of the population. The abundance of Actinobacteria was similar (15%) in both the regions.
Figure 4: Percentage of phylogenetic groups in OMZ and non-OMZ sites.
Discussion
The sampling stations in the present study form a spatial gradient along the continental margin. The site at 50m is located in the continental shelf while the sites at 200m, 500m and 1000m are situated in the continental slope. In this region there is a vertical decrease in the dissolved oxygen concentrations from 0.5mlL-1 to as low as 0mlL-1. On the other hand, the site in the continental shelf is well ventilated with dissolved oxygen concentrations 3-5mlL-1. Oxygen is a strong evolutionary force and the dominating factor determining functional interactions and spatial structure of many microbial communities [12]. Though studies have been carried out on the diversity of bacteria in the OMZ water systematic studies on the bacterial diversity in the sediments is very low [12-16]. Few studies have been conducted in the OMZ sediments of Pacific and Atlantic Ocean [17,18]. Bacterial diversity of the Arabian Sea OMZ water column using cultivation-independent methods have been conducted [13,14,16], but those in Arabian Sea OMZ sediments is meagre [19]. This is the first report of the bacterial diversity in the surficial sediments using PCR-DGGE analysis.
The important groups obtained from this study were Proteobacteria, Acidobacteria, Actinobacteria, Chloroflexi, Nitrospira and Bacteriodetes. Proteobacteria was the dominant group observed in all sites and transects. Proteobacteria is ubiquitous in distribution and their dominance has been reported in AS OMZ water [16]. The Deltaproteobacteria, which includes the anaerobic sulphate reducing bacteria, were the most abundant class, suggesting the prevalence of sulphate reduction in this region. Sulphate reducing bacteria falling in genera Desulfosarcina and Desulfofrigus have been retrieved from the OMZ waters of Arabian Sea [13,20]. 73% of the sequences showed close affiliation to Syntrophobacterales and Desulfovibrionales. Sulphate concentrations varied from 39.8 to 12.7mm in the pore waters of the eastern AS indicating sulphate reduction [21]. Similar occurrence of high sulphate reducers has been reported in other marine sediments [22]. The dominance of this group was not restricted only to Mid Atlantic Ridge [23] but also in Tokyo and Sagami bays [24,25]. However, a few did not show any resemblance to cultured representatives but were closely related to clones detected at cold seeps and mud volcano [26,27].
Gammaproteobacteria formed a major group after Deltaproteobacteria in the phylum Proteobacteria in the both the sites with higher occurrence in the OMZ. Occurrence of this has been reported among culturable and non-culturable bacteria of pelagic waters and sediments [28,29]. There have also been reports of active diastrophic planktonic Gammaproteobacteria in the Arabian Sea [30]. Thus the possibilities of the presence of sediment diazotrophic gammaproteobacterial communities cannot be ruled out and future studies should be conducted using functional probes. Also the largest bacteria which have been reported such as Thioploca sp. falling in the Gammaproteobacteria in the OMZ sediments off Oman have not been observed in OMZ sediments in the present study [31]. Actinobacteria have been found both in the photic zone and in deep water, and seem to be more frequently recovered in deep water [32]. The presence of Actinobacteria in marine ecosystems has been attributed by some to run off from fringing terrestrial habitat [33]. Other studies have reported a widespread and persistent occurrence of indigenous actinobacterial populations in marine sediments due to their physiological adaptations for growth in the marine environments [34,35]. These results suggest that Actinobacteria are adapted to marine environment and may be a dominant group in this ecosystem as they are in soils.
Chloroflexi was restricted to the deeper two OMZ sites. The presence of Chloroflexi related SAR 202 cluster, which forms a monophyletic subgroup in the phylum, has been reported in the oxic and the oxygen minimum zone waters although these were not found in the present study [14,36]. The members of this phylum exhibit unusual metabolic diversity like anoxygenic photosynthesis, organotrophy and reduction of chlorinated hydrocarbons [37]. Schafer et al. [18] in their work on the microbial diversity of the sediment of Benguela upwelling system reported the presence of this phylum in sulphate-depleted depths [18]. Their presence seems to be a common feature of many deep sub- seafloor sediments [38,39]. Thus the present studies demonstrate that eastern AS OMZ region possesses a phylogenetically distinct bacterial community that may be playing an important role in the prevailing biogeochemical processes. The data obtained from DGGE profiling is more accurately considered as the structure of dominant populations rather than a general measure of bacterial diversity [40]. The deeper depths i.e., OMZ region lodged more bacterial populations than non-OMZ region suggesting that the environmental conditions prevalent in these regions provide a favorable condition for the survival of bacteria.
Acknowledgement
We are grateful to the captain and crew of ORV Sagar Sampada for the collection of samples. This work was supported by the grant in aid from Central Marine Living Resources and Ecology (CMLRE), Kochi and laboratory facilities of Marine Microbial Reference Facility of NIO RC, Kochi funded by the Ministry of Earth Sciences, New Delhi. BD and AF thank Council of Scientific and Industrial Research, New Delhi, for the award of Junior and Senior Research Fellowship.
References
- Kamykowski D, Zentara SJ (1990) Hypoxia in the world ocean as recorded in the historical data set. Deep Sea Research Part A oceanographic Research Papers 37(12): 1861-1874.
- Stramma L, Johnson GS, Sprintall J, Mohrholz V (2008) Expanding oxygen-minimum zones in the tropical oceans. Science 320(5876): 655658.
- Keeling RF, Kortzinger A, Gruber N (2010) Ocean deoxygenation in a warming world. Ann Rev in Mar Sci 2: 199-229.
- Helly JJ and Levin LA (2004) Global distribution of naturally occurring marine hypoxia on continental margins. Deep Sea Research Part I oceanographic Research Papers 51(9): 1159-1168.
- Berner RA (1989) Biogeochemical cycles of carbon and sulphur and their effect on atmospheric oxygen over Phanerozoic time. Paleogeography Paleoclimate & Paleoecology 75(1-2): 97-122.
- Hermelin JOR (1992) Variations in the benthic foraminiferal fauna of the Arabian Sea: a response to changes in upwelling intensity? Geological Society Special Publication 64: 151-166.
- Ducklow HW, Carlson CA (1992) Oceanic bacterial production. In: Marshall KC (Ed.), Advances in Microbial Ecology Plenum. Springer, USA, vol 12, pp. 113-181.
- Ward DM, Weller R, Bateson MM (1990) 16SrRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 345(6270): 63-65.
- Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S ribosomal RNA. Appl Environ Microbiol 59(3): 695-700.
- Zhou JZ, Bruns MA, Tiedje JM (1996) DNA recovery from soils of diverse composition. Applied and Environmental Microbiology 62(2): 316-322.
- Kumar S, Tamura K, Nei M (2004) MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5(2): 150-163.
- Fenchel T, Finlay B (2008) Oxygen and the spatial structure of microbial communities. Biol Rev Camb Philos Soc 83(4): 553-569.
- Fuchs BM, Woebken D, Zubkov MV , Burkill P, Amann R, et al. (2005) Molecular identification of picoplankton populations in contrasting waters of the Arabian Sea. Aquatic Microbial Ecology 39(2): 145-157.
- Stevens H, Ulloa O (2008) Bacterial diversity in the oxygen minimum zone of the eastern tropical South Pacific. Environmental Microbiology 10(5): 1244-1259.
- Molina V, Farias L (2009) Aerobic ammonium oxidation in the oxycline and oxygen minimum zone of the eastern tropical South Pacific off northern Chile (~20 °S). Deep Sea Research Part II Topical Studies in Oceanography 56(16): 1032-1041.
- Jain A, Bandekar M, Gomes J, Shenoy D, Meena RM, et al. (2014) Temporally invariable bacterial community structure in the Arabian Sea oxygen minimum zone. Aquatic Microbial Ecology 73(1): 51-67.
- Liu X, Tiquia T, Holguin G , Wu L, Nold SC, et al. (2003) Molecular diversity of denitrifying genes in continental margin sediments within the oxygen deficient zone off the Pacific coast of Mexico. Applied and Environmental Microbiology 69(6): 3549-3560.
- Schafer H, Ferdelman TG, Fossing H, Muyzer G (2007) Microbial diversity in deep sediments of the benguela upwelling system. Aquatic Microbial Ecology 50(1): 1-9.
- Divya B, Parvathi A, Loka Bharathi PA, Nair S (2011) 16S rRNA- based bacterial diversity in the organic-rich sediments underlying oxygen-deficient waters of the eastern Arabian Sea. World Journal of Microbiology & Biotechnology 27(12): 2821-2833.
- Riemann L, Steward GF, Fandino LB, Campbell L , Landry MR, et al. (1999) Bacterial community composition during two consecutive NE Monsoon periods in the Arabian Sea studied by denaturing gradient gel electrophoresis (DGGE) of rRNA genes. Deep Sea Research Part II : Topical Studies in Oceanography 46(8-9): 1791-1811.
- Karisiddaiah SM, Borole DV, Rao BR, Paropkari AR, Joao HM, et al. (2006) Studies on the pore water sulphate, chloride and sedimentary methane to understand the sulphate reduction process in eastern Arabian Sea. Current Science 91(1): 82-86.
- Gray JP, Herwig RP (1996) Phylogenetic analysis of the bacterial communities in marine sediments. Applied and Environmental Microbiology 62(11): 4049-4059.
- Lopez-Garcia P, Duperron S , Philippot P , Foriel J , Susini J, et al. (2003) Bacterial diversity in hydrothermal sediment and epsilon proteobacterial dominance in experimental microcolonizers at the mid Atlantic Ridge. Environmental Microbiology 5(10): 961-976.
- Urakawa H, Kita-Tsukamoto K, Ohwada K (1999) Microbial diversity in marine sediments from Sagami Bay and Tokyo Bay, Japan, as determined by 16S rRNA gene analysis. Microbiology 145(Pt 11): 3305-3315.
- Urakawa H, Yoshida T, Nishimura M, Ohwada K (2000) Characterization of depth related population variation in microbial communities of a coastal marine sediment using 16S rDNA based approaches and quinone profiling. Environmental Microbiology 2(5): 542-554.
- Fang JS, Shizuka A, Kato C, Schouten S (2006) Microbial diversity of cold seep sediments in Sagami Bay, Japan, as determined by 16S rRNA gene and lipid analyses. FEMS Microbiology Ecology 57(3): 429-441.
- Losekann T, Knittel K, Nadalig T, Fuchs B, Niemann H, et al. (2007) Diversity and abundance of aerobic and anaerobic methane oxidizers at the Haakon Mosby mud volcano, Barents Sea. Appl Environ Microbiol 73(10): 3348-3362.
- Lopez-Garcia P, Lopez-Lopez A, Moreira D, Rodriguez-Valera F (2001) Diversity of free-living prokaryotes from a deep sea site at the Antarctic Polar Front. FEMS Microbiol Ecol 36(2-3): 193-202.
- Cho JC, Giovannoni SJ (2004) Cultivation and growth characteristics of a diverse group of oligotrophic marine Gammaproteobacteria. Applied and Environmental Microbiology 70(1): 432-440.
- Bird C, Martinez JM, O'Donnell AG, Wyman M (2005) Spatial distribution and transcriptional activity of an uncultured clade of planktonic diazotrophic gammaproteobacteria in the Arabian Sea. Appl Environ Microbiol 71(4): 2079-2085.
- Schmaljohann R, Drews M, Walter S, Linke P, von Rad C, et al. (2001) Oxygen-minimum zone sediments in the northeastern Arabian Sea off Pakistan: a habitat for the bacterium Thioploca. Marine Ecology Progression Series 211: 27-42.
- DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, et al. (2006) Community genomics among stratified microbial assemblages in the ocean's interior. Science 311(5760): 496-503.
- Piza FF, Prado PI, Manfio GP (2004) Investigation of bacterial diversity in Brazilian tropical estuarine sediments reveals high actinobacterial diversity. Antonie van Leeuwenhoek 86(4): 317-328.
- Colquhoun JA, Mexson J, Goodfellow M, Ward AC, Horikoshi K, et al. (1998) Novel rhodococci and other mycolate actinomycetes from the deep sea. Antonie van Leeuwenhoek 74(1-3): 27-40.
- Mincer TJ, Jensen PR, Kauffman CA, Fenical W (2002) Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl Environ Microbiol 68(10): 5005-5011.
- Morris RM, Rappe MS, Urbach E, Connon SA, Giovannoni SJ (2004) Prevalence of the Chloroflexi-related SAR202 bacterioplankton cluster throughout the mesopelagic zone and deep ocean. Appl Environ Microbiol 70(5): 2836-2842.
- Maymo-Gatell X, Tandoi V, Gossett JM, Zinder SH (1995) Characterization of an H2 utilizing enrichment culture that reductively dechlorinates tetrachloroethene to vinyl chloride and ethene in the absence of methanogenesis and acetogenesis. Appl Environ Microbiol 61(11): 3928-3933.
- Jorgensen BB, D'Hondt S (2006) Ecology - A starving majority deep beneath the seafloor. Science 314(5801): 932-934.
- Teske AP(2006) Microbial communities of deep marine subsurface sediments: molecular and cultivation surveys. Geomicrobiology Journal 23(6): 357-368.
- Kowalchuk GA, Drigo B, Yergeau E, Veen JA (2006) Assessing bacterial and fungal community structure in soil using ribosomal RNA and other structural gene markers. Nucleic acids and proteins in soil 8: 159-188.
© 2017 Baby Divya, et al. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






