- Submissions

Full Text
Cohesive Journal of Microbiology & Infectious Disease
Comparative Molecular Evolution of the Most Dominant and Virulent Recombinant Sars-CoV-2 Variant XE and its Parent Lineage Viruses BD.1 and BA. 2.30
Meliscan Arlı1 and Efe Sezgin1,2*
1Department of Biotechnology, Izmir Institute of Technology, Turkey
2Department of Food Engineering, Izmir Institute of Technology, Turkey
*Corresponding author: Efe Sezgin, Department of Food Engineering, Izmir Institute of Technology, Turkey
Submission: May 09, 2025;Published: October 09, 2025

ISSN 2578-0190 Volume7 issues 5
Abstract
Sars-CoV-2 is a globally circulating virus chronically infecting tens of millions of people every year. In addition to individual mutations, with the advance of the Omicron variant, novel recombinant viruses are emerging. Similar to the effect of mutations on the infectivity and disease dynamics, recombination has the potential to create dangerous viruses. Despite many studies on the mutation-driven variants of Sars- CoV-2, the effect of recombination on the molecular evolution of the viruses is not known. We aimed to compare the molecular evolution of the most dominant and virulent recombinant Sars-CoV-2 variant XE and its parent lineage viruses BD.1 and BA.2.30 using population genetics and phylogenetic methods. High coverage full genome sequences of XE, BD.1 and BA.2.30 viruses were obtained from the Global Initiative on Sharing All Influenza Data (GISAID) web portal. Sequence alignments and phylogenetic analyses were performed by Multiple Alignment using Fast Fourier Transform (MAFFT) and Molecular Evolutionary Genetic Analysis (MEGA), respectively. Annotation was performed using whole genome of the Wuhan type Sars-CoV-2 virus. Population genetic tests included nucleotide diversity, diversification, selection and demographic change tests. Effect of amino acid changes on protein structure and functionality was investigated. Results suggest that recombinant XE variant emerged as a recombination between BD.1 and BA.2.30 sequences most probably by late 2021/early 2022 in Europe and disappeared by the end of 2022. XE genomes had different molecular evolutionary dynamics compared to its parental lineages. The molecular evolution of XE sequences were driven by high number of rare/singleton non-synonymous changes acquired after the recombination event, most of which lead to non-conservative amino acid changes that were eliminated from population level circulation rather quickly. Effective neutralization by host immune system and lack of adaptive mutations that could have conferred increased transmissibility and immune evasion to XE might have led to the eradication of XE.
Keywords: Sars-CoV-2; Recombination; XE variant; Molecular evolution; Selection
Introduction
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), the virus responsible for Coronavirus Disease 2019 (COVID-19), was first reported in late 2019 from Wuhan, China [1]. Since being identified, SARS-CoV-2 has spread across the globe resulting in over hundreds of millions of cases of COVID-19 and over seven million deaths [2]. Despite vaccination and other protective measures SARS-CoV-2 is circulating across the globe causing persistent infections that inflict severe negative impact on global health [3,4].
SARS-CoV-2 has a single stranded positive-sense RNA genome around 30 kilo bases that codes for 16 Non-Structural Proteins (NSPS), 4 structural proteins and 9 accessory proteins [5]. Similar to other RNA viruses Sars-CoV-2 evolve rapidly driven by high mutation rate [6,7]. Mutations drive evolution of new lineages and affect viral phenotypes such as transmissibility, onset of infectiousness, duration of infectiousness and immune evasion [8-14]. In addition to individual mutations, recombination is another mechanism that generates viral genetic diversity. Sars-CoV-2 recombination is the process where a host is infected with at least two genetically distinct viruses (or lineages); these viruses recombine within the host and produce viable progeny that can spread to other hosts [15- 17]. The XA lineage (designated by the Pango nomenclature) was the first reported Sars-CoV-2 recombinant lineage [18]. Since then, several other recombinant lineages have been reported [16,19- 22]. Recombination has the potential to bring genetic backgrounds with different viral phenotypes together creating novel viral phenotypes with enhanced infectivity, transmissibility and immune evasion [23]. Therefore, continuing surveillance and studies on the molecular evolution of recombinant strains is crucial.
Although there are many studies on the mutation-driven variants of Sars-CoV-2, the effect of recombination on the molecular evolution of the viruses is not known. We aimed to compare and contrast the molecular evolution of one of the most dominant and virulent recombinant Sars-CoV-2 variants XE and its parent lineage viruses BD.1 and BA.2.30. Our analyses could provide insights on the evolutionary forces acting on recombinant viral genomes and how these processes shape viral phenotypes such as transmissibility and immune escape.
Materials and Methods
All XE, BA.2.30 and BD.1 whole genome sequences analysed in this study were downloaded from GISAID (https://gisaid.org/) and can be reached at EPI_SET_240818dm (2960 XE, BA.2.30 and BD.1 genome sequences submitted in 2022, doi: 10.55876/ gis8.240818dm) and EPI_SET_240811dv (139 high quality full XE, BA.2.30 and BD.1 genome sequences submitted in 2022 used in molecular population genetic analyses, doi: 0.55876/ gis8.240811dv). The Wuhan-type reference Sars-CoV-2 genome and genome annotation was downloaded from NCBI (https:// www.ncbi.nlm.nih.gov/nuccore/1798174254). Genome sequences of at least 29 Kilo Bases (kb) long, with less than 1% unknown nucleotide assignment (i.e. N rather than A, T, C, G nucleotides), with less than 0.05% unique mutations and with no unconfirmed insertion/deletion mutations were used for analyses.
The sequences were aligned using the Muscle algorithm in MAFFT (v7.520) [24]. Annotation was performed using whole genome of the Wuhan-type virus. Visual inspection of sequence alignments and phylogenetic analyses were conducted in MEGA11 (https://www.megasoftware.net/) [25]. Molecular population genetics analyses included estimation of the number of mutations per site (Eta), segregating Sites (S), average number of nucleotide diversity between two sequences (represented by π [26,27] and Watterson estimator of proportion of polymorphic sites (represented by θ) [28]. Selection and demographic change analyses included Tajima F [29], Fu-Li’s D* and Fu YX et al. [30] tests. All population genetics tests were conducted in DnaSP 6 [31].
The nature of amino acid changes and potential effect on protein structure were determined by Expasy online tools (https:// www.expasy.org/), Jalview [32], R (https://www.R-project.org/) packages Biostrings (https://bioconductor.org/packages/release/ bioc/html/Biostrings.html) and seqinr (https://seqinr.r-forge.rproject. org/).
Results and Discussion
Origin and temporospatial distribution of XE sequences
XE, BD.1 and BA.2.30 whole genome sequence alignment analyses showed that all XE sequences are products of recombination between parental BD.1 and BA.2.30 sequences that happened within the Nsp 6 gene region (Figure 1a). The regions between the Nsp 1-Nsp 6 and Nsp 7-Orf 10 genes of the XE lineage genomes represent BD.1 and BA.2.30 background, respectively (Figure 1a). Our findings are in agreement with the previous reports [16,23].
In total 2960 XE sequences were found on the GISAID database all reported in the year 2022. There were only 12 sequences reported in January 2022 from Europe. The highest number of XE sequences were reported in March and April 2022. No XE sequences were reported at GISAID past October 2022 (Figure 1b). Eighty-one percent (2412/2960) of the sequences were reported from Europe. XE sequences outside of Europe (Asia, North and South America, Oceania) were reported after March 2022. These observations suggest that the first XE recombinant lineages emerged at the end of 2021 or January 2022 in Europe, spread around the globe, but disappeared by the end of 2022. Phylogenetic analysis of representative XE, BD.1 and BA.2.30 whole genome sequences sampled through different months in 2022 showed that XE sequences form a distinct clade separate from parental BD.1 and BA.2.30 sequences (Figure 1c).
Figure 1: The recombination point on the Sars-CoV-2 genome that created the XE recombinant variant from the parental BA.2.30 and BD.1 variants together with Sars-CoV-2 genome annotation (A). Monthly distribution of recombinant XE variant sequences reported on the GISAID (https://gisaid.org/) web site (B). Maximum-likelihood phylogenetic tree of BA.2.30, BD.1 and XE whole genome sequences (C). There are 10 branches in each of the BA.2.30, BD.1 and XE clades representing one full genome sequence sampled from January (top branch) to October (bottom branch) 2022.

Comparative molecular evolution of XE, BD.1 and BA.2.30 sequences
In order to test whether XE viruses had different molecular evolution dynamics compared to the parental BD.1 and BA.2.30 viruses we conducted molecular population genetics analyses. Genome-wide sliding window analysis of overall nucleotide diversity showed that the nucleotide diversity of XE sequences was different compared to parental BD.1 and BA.2.30 sequences, particularly being higher at the recombination region, driven by higher number of rare/singleton mutations (Figure 2a) (Table 1). Genome-wide sliding window analyses of Fu-Li’s D and Tajima’s D tests with their rather negative results also supported excess of rare/singleton variants in XE genomes compared to parental BD.1 and BA.2.30 sequences (Figure 2b & 2c). Fu-Li’s D test examines the distribution of new and old mutations on a sequence/gene tree. Negative Fu-Li’s D test results indicate that most mutations are on the outer branches of a tree and thus they are new mutations and happened after the recombination event. Exponential population growth, negative (including background) selection or action of both of these factors can lead to negative Tajima’s D values. To uncover which of these evolutionary forces are the dominant factor driving the XE sequence evolution, we conducted additional Tajima’s D tests separately for the synonymous and non-synonymous changes on the whole genome sequence and the Nsp 1-Nsp 6 (BD.1 background) and Nsp 7-ORF10 (BA.2.30 background) genome regions of XE. Then, we conducted the same tests on the same genome regions for BD.1 and BA.2.30 and compared the results. Tajima’s D results for the synonymous and non-synonymous changes were more negative for XE sequences compared to BD.1 and BA.2.30 sequences (Table 1). Moreover, the non-synonymous Tajima’s D values were even more negative than the synonymous Tajima’s D results (Table 1).
Figure 2: Sliding window analyses of (A) nucleotide diversity, (B) Fu and Li’s D and (C) Tajima’s D tests along the BA.2.30, BD.1 and recombinant XE genomes.

Table 1:Population genetics summary statistics and tests comparing recombinant XE and parental BD.1 and BA.2.30 sequences.

Abbreviations: S: Number of segragating sites; Eta: total number of mutations; Singleton: Number of singleton changes; Syn. Pol: Number of synonymous changes; Rep. Pol: Number of Replecement (non-synonymous) changes; Theta-W: Watterson theta nucleotide diversity estimate; π Syn. Sites: Pairwise nucleotide diversity of only synonymous sites; π Non-syn. Sites: Pairwise nucleotide diversity of only non-synonymous sites; TD: Tajima’s D; TD Syn: Tajima’s D calculation for synonymous sites; TD Non-syn: Tajima’s D calculation for non-synonymous sites.
Theta-W, π All Sites, π Syn. Sites and π Non-syn. Site values should be multiplied by 10-4.
Number of sequences analyzed: XE n=66; BD.1 n=33; BA.2.30 n=20.
*: p<0.05; **: p< 0.01; ***: p<0.001.
Divergence between the parental lineages and XE sequences were calculated by dN/dS ratios which is the ratio of nonsynonymous mutations per non-synonymous site (dN) to synonymous mutations per synonymous site (dS). The dN/dS ratio of XE and BD.1 sequences for Nsp 1-Nsp 6 (BD.1 background) regions was 1.03. The dN/dS ratio of XE and BA.2.30 sequences for Nsp 7-ORF10 (BA.2.30 background) regions was 0.89. The whole genome dN/dS ratio between BD.1 and BA.2.30 was 0.48. These results show that there is increased non-synonymous site divergence between XE and parental BD.1 and BA.2.30 sequences.
Examination of the nature (transitions and transversions) of nucleotide changes showed that C > T transition was the most common mutation observed in both synonymous and nonsynonymous changes in XE, BD.1 and BA.2.30 genomes (Table 2). C > T transitions were responsible for around 31% of nonsynonymous changes in XE, BD.1 and BA.2.30 genomes (Table 2). In terms of synonymous changes, C > T transitions accounted for 72%, 32% and 67% of all synonymous changes in XE, BD.1 and BA.2.30 genomes, respectively (Table 2). Human APOBEC is reported to edit viral genomes in excess of C > T transition [33-35], which may explain the dominance of C > T transitions in XE, BD.1 and BA.2.30 genomes. A > G and G > A transitions were the next abundant transitions observed in XE, BD.1 and BA.2.30 genomes (Table 2). Human antiviral protein ADAR1 could be responsible for increased A > G transitions in these viral genomes [35,36]. The results for transition and transversion analyses suggest that human antiviral protein activity may be an important mutation driver in XE, BD.1 and BA.2.30 genomes.
Table 2:Transition and transversion nature of nucleotide changes observed in XE, BD.1 and BA.2.30 genomes.
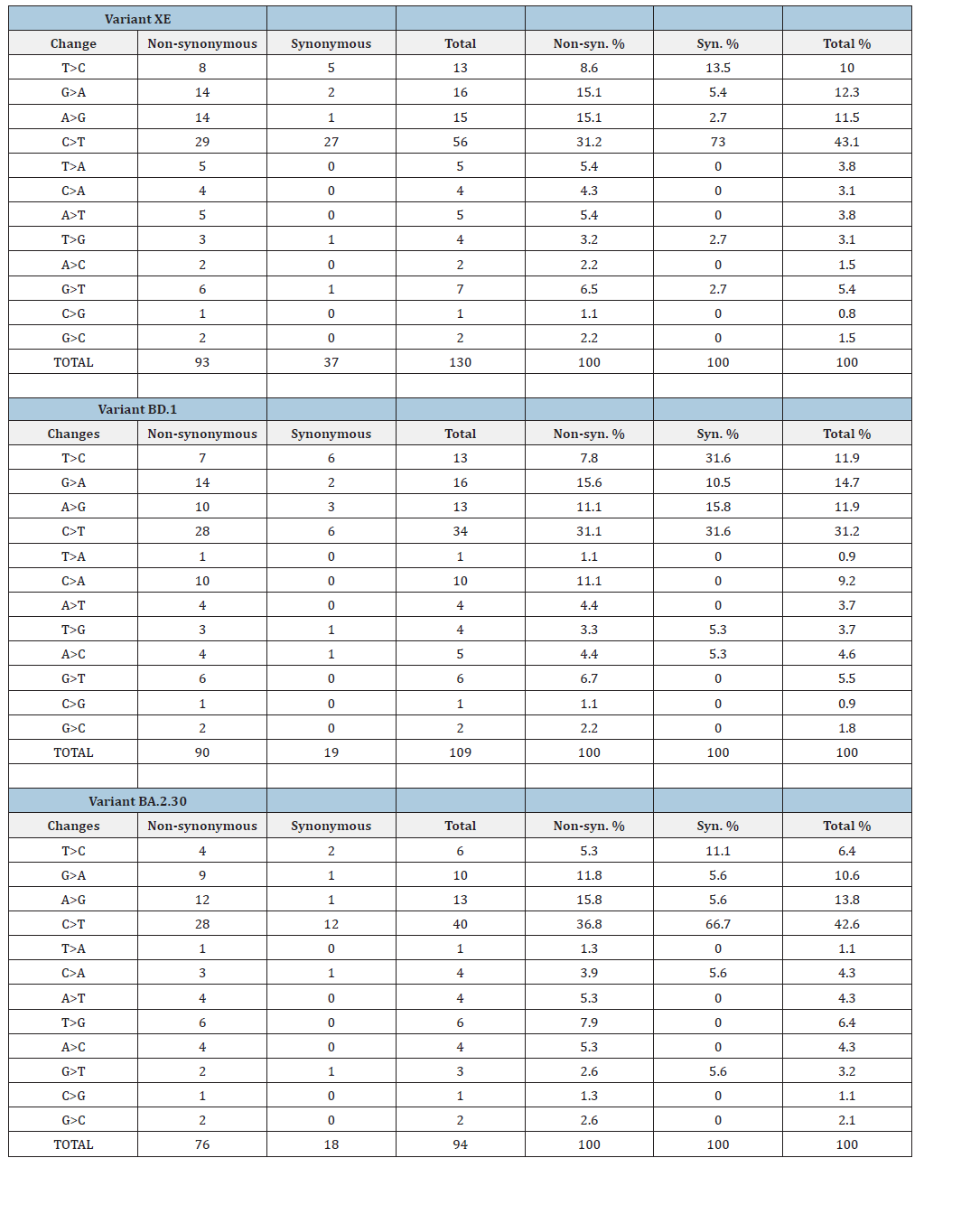
Considering the total number of segregating sites (S) and number of mutations (Eta) throughout the XE genome, 71% (60/84) were singletons and the number of replacement (non-synonymous) changes were 1.5 times more (50 vs 34) than synonymous changes. Very similar trends were observed for BD.1 and BA.2.30 genomes. Comparison of same analyses with Nsp 1-Nsp 6 (BD.1 background) and Nsp 7-ORF10 (BA.2.30 background) genome regions of XE to the same BD.1 and BA.2.30 genome regions also yielded very similar results. These results suggest that similar to the molecular evolution dynamics of its parental lineages BD.1 and BA.2.30, more replacement mutations than silent (synonymous) mutations occur on XE genomes; however, negative selection eliminates or keeps these possibly damaging replacement mutations at a very low frequency. This suggestion is also supported by the comparisons of the nucleotide diversity estimates of silent and replacement sites. Considering the whole genome and the Nsp 1-Nsp 6 (BD.1 background) and Nsp 7-ORF10 (BA.2.30 background) genome regions the nucleotide diversity of synonymous sites was higher than that of non-synonymous sites despite much higher number of non-synonymous changes observed in these genome regions (Table 1). Purifying (negative) selection should be eliminating non-synonymous changes leading to lower non-synonymous site nucleotide diversity.
Examination of the nature of total replacement changes observed on XE, BD.1 and BA.2.30 genomes showed that 80 of the amino acid changes were conservative whereas 173 were nonconservative (such as leading the charge, polarity, size changes that may affect protein structure) changes (Table 3). When examined separately, the number of non-conservative changes were nearly double the number of conservative changes for XE, BD.1 and BA.2.30 genomes (Table 3). Highest number of non-conservative changes were observed in the Spike gene. We also examined the longitudinal propagation of replacement changes observed on XE genomes. None of the replacement changes survived past two months. For example, a replacement changes first reported in March 2022 was not observed in the XE genome sequences reported past April 2022 indicating that these changes disappeared within a few months’ time. This supports the hypothesis that purifying (negative) selection eliminates possibly deleterious mutations. Nevertheless, one cannot rule out the fact that at least some of these mutations were lost due to transmission bottleneck and genetic drift [37].
Table 3:The number of amino acid changes observed in XE, BD.1 and BA.2.30 genomes. Conservative and nonconservative changes are listed under separate columns.

Since rather high numbers of synonymous and non-synonymous changes were observed on XE genomes, we investigated whether these changes were inherited from the parental (BD.1 and BA.2.30) genomes (already present before the recombination) or happened after the recombination event and were unique to the XE genomes. We compared the number and distribution of XE-specific and XE-non-specific synonymous and non-synonymous changes throughout the XE genomes. Considering the whole genome, the number of XE-specific (changes only seen on the XE genomes not on the BD.1 and BA.2.30 genomes) synonymous and non-synonymous changes were higher than XE-non-specific (changes also seen either on the BD.1 and BA.2.30 genomes) synonymous and nonsynonymous changes (Figure 3a). For the Nsp 1-Nsp 6 (BD.1 background) genome region the number of XE-specific synonymous and non-synonymous changes were much higher than XE-nonspecific (changes also seen on the BD.1 genomes) changes (Figure 3b), whereas for the Nsp 7-ORF10 (BA.2.30 background) genome region the number of XE-specific and XE-non-specific (changes also seen on the BA.2.30 genomes) synonymous and non-synonymous changes were similar (Figure 3c). These results show that the number of mutations unique to XE genomes (happened after the recombination event) are much higher than mutations inherited from the parental genomes suggesting increased mutation rate, at least on the BD.1 genome background region, on the XE genomes.
Figure 3: Comparisons of the number of XE lineage-specific versus XE lineage-non-specific synonymous and nonsynonymous mutations considering (A) whole genome, (B) genome region between Nsp 1 and Nsp 6 representing the BD.1 genomic background, (C) genome region between Nsp 7 and ORF10 representing the BA.2.30 genomic background and (D) only the Spike gene. XE lineage-specific mutations represent mutations that happened after the recombination event. XE lineage-non-specific mutations are the mutations that are also observed on the parental lineages.
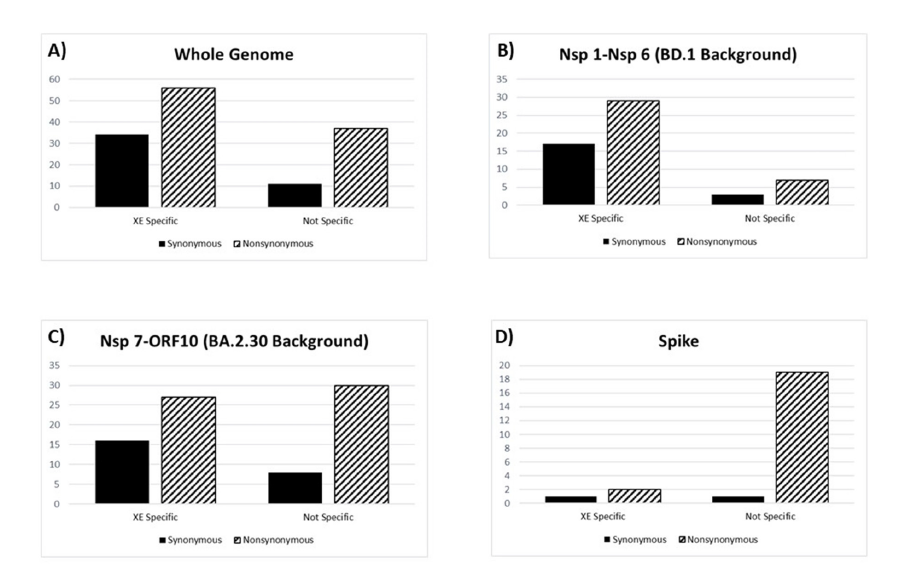
Finally, we compared the molecular evolution of Spike gene among XE, BD.1 and BA.2.30 lineages. All population genetics parameter estimates (such as the number of segregating sites, nucleotide diversity, Tajima’s D, and Fu-Li tests) were very different between BD.1 and BA.2.30 samples (Table 1). As the Spike gene is in the BA.2.30 background region in the XE genome, we expected the Spike gene parameter estimates of the XE samples to be more similar to BA.2.30 samples. Indeed, the parameter estimates of XE and BA.2.30 Spike genes were similar; however, more singleton synonymous and non-synonymous changes were observed for XE Spike gene (Table 1). Analysis of XE-specific synonymous and nonsynonymous changes in the Spike gene showed that most of the synonymous and non-synonymous changes were not XE-specific; thus, they were already present in the BA.2.30 background. This pattern is in contrast with the rest of the genome where most mutations were XE specific.
Conclusion
Recombinant XE variant emerged as a recombination between BD.1 and BA.2.30 sequences most probably by late 2021/early 2022 in Europe and disappeared by the end of 2022. XE genomes had different molecular evolutionary dynamics compared to its parental lineages. The molecular evolution of XE sequences were driven by high number of rare/singleton non-synonymous changes acquired after the recombination event, most of which lead to non-conservative amino acid changes that were eliminated from population level circulation rather quickly. Unlike the rest of the genome the XE Spike gene was rather similar to parental BA.2.30 Spike gene that might have helped XE to spread. On the other hand, conservation of spike antibody binding regions might have enabled the host immune system to effectively neutralize XE since the host immune systems were already primed by the parental BA.2.30 viruses. In addition, lack of adaptive mutations that could have conferred increased transmissibility and immune evasion to XE might have led to the eradication of XE.
Conflict of Interest Statement
Authors declare no competing interests.
References
- Burki T (2020) The origin of SARS-CoV-2. Lancet Infect Dis 20(9): 1018-1019.
- World Health Organization (2024) World health organization COVID-19 dashboard. Number of COVID-19 deaths reported to WHO (cumulative total).
- Tracking SARS-CoV-2 variants.
- SARS-CoV-2 variants of concern as of 26 July 2024.
- Gorkhali R, Koirala P, Rijal S, Mainali A, Baral A, et al. (2021) Structure and function of major SARS-CoV-2 and SARS-CoV proteins. Bioinform Biol Insights 15: 11779322211025876.
- Amicone M, Borges V, Alves MJ, Isidro J, Zé-Zé L, et al. (2022) Mutation rate of SARS-CoV-2 and emergence of mutators during experimental evolution. Evol Med Public Health 10(1): 142-155.
- Dorp LV, Acman M, Richard D, Shaw LP, Ford CE, et al. (2020) Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect Genet Evol 83: 104351.
- Sinha S, Tam B, Wang SM (2021) RBD double mutations of SARS-CoV-2 strains increase transmissibility through enhanced interaction between RBD and ACE2 receptor. Viruses 14(1): 1.
- Ozono S, Zhang Y, Ode H, Sano K, Tan TS, et al. (2021) SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat Commun 12(1): 848.
- Thorne LG, Bouhaddou M, Reuschl AK, Zuliani-Alvarez L, Polacco B, et al. (2022) Evolution of enhanced innate immune evasion by SARS-CoV-2. Nature 602(7897): 487-495.
- Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, et al. (2021) SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 19(7): 409-424.
- Carabelli AM, Peacock TP, Thorne LG, Harvey WT, Hughes J, et al. (2023) SARS-CoV-2 variant biology: Immune escape, transmission and fitness. Nat Rev Microbiol 21(3): 162-177.
- Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, et al. (2021) Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe 29(1): 44-57.
- Obermeyer F, Jankowiak M, Barkas N, Schaffner SF, Pyle JD, et al. (2022) Analysis of 6.4 million SARS-CoV-2 genomes identifies mutations associated with fitness. Science 376(6599): 1327-1332.
- Jackson B, Boni MF, Bull MJ, Colleran A, Colquhoun RM, et al. (2021) Generation and transmission of interlineage recombinants in the SARS-CoV-2 pandemic. Cell 184(20): 5179-5188.
- Focosi D, Maggi F (2022) Recombination in coronaviruses, with a focus on SARS-CoV-2. Viruses 14(6): 1239.
- Pollett S, Conte MA, Sanborn M, Jarman RG, Lidl GM, et al. (2021) A comparative recombination analysis of human coronaviruses and implications for the SARS-CoV-2 pandemic. Sci Rep 11(1): 17365.
- O’Toole Á, Hill V, Pybus OG, Watts A, Bogoch II, et al. (2021) Tracking the international spread of SARS-CoV-2 lineages B.1.1.7 and B.1.351/501Y-V2 with grinch. Wellcome Open Res 6: 121.
- Sekizuka T, Itokawa K, Saito M, Shimatani M, Matsuyama S, et al. (2022) Genome recombination between the delta and alpha variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Jpn J Infect Dis 75(4): 415-418.
- Gutierrez B, Sánchez HG, Candido D, Jackson B, Fleishon S, et al. (2022) Emergence and widespread circulation of a recombinant SARS-CoV-2 lineage in North America. Cell Host Microbe 30(8): 1112-1123.
- Silva T, Salvato RS, Gregianini TS, Gomes IA, Pereira EC, et al. (2022) Molecular characterization of a new SARS-CoV-2 recombinant cluster XAG identified in Brazil. Front Med 9: 1008600.
- Scarpa F, Locci C, Azzena I, Casu M, Fiori PL, et al. (2023) SARS-CoV-2 recombinants: Genomic comparison between XBF and its parental lineages. Microorganisms 11(7): 18-24.
- Shiraz R, Tripathi S (2023) Enhanced recombination among omicron subvariants of SARS-CoV-2 contributes to viral immune escape. J Med Virol 95(2): e28519.
- Katoh K, Rozewicki J, Yamada KD (2019) MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20(4): 1160-1166.
- Tamura K, Stecher G, Kumar S (2021) MEGA11: Molecular evolutionary genetics analysis version 11. Mol Biol Evol 38(7): 3022-3027.
- Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76(10): 5269-73.
- Nei M, Tajima F (1981) DNA Polymorphism detectable by restriction endonucleases. Genetics 97(1): 145-163.
- Watterson GA (1975) On the number of segregating sites in genetical models without recombination. Theor Popul Biol 7(2): 256-276.
- Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123(3): 585-595.
- Fu YX, Li WH (1993) Statistical tests of neutrality of mutations. Genetics 133(3): 693-709.
- Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, et al. (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol 34(12): 3299-3302.
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009) Jalview version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 25(9): 1189-1191.
- Klimczak LJ, Randall TA, Saini N, Li JL, Gordenin DA (2020) Similarity between mutation spectra in hypermutated genomes of rubella virus and in SARS-CoV-2 genomes accumulated during the COVID-19 pandemic. PLoS One 15(10): e0237689.
- Simmonds P (2020) Rampant C→U hypermutation in the genomes of SARS-CoV-2 and other coronaviruses: Causes and consequences for their short- and long-term evolutionary trajectories. mSphere 5(3): e00408-20.
- Giorgio S, Martignano F, Torcia MG, Mattiuz G, Conticello SG (2020) Evidence for host-dependent RNA editing in the transcriptome of SARS-CoV-2. Sci Adv 6(25): eabb5813.
- Ringlander J, Fingal J, Kann H, Prakash K, Rydell G, et al. (2022) Impact of ADAR-induced editing of minor viral RNA populations on replication and transmission of SARS-CoV-2. Proc Natl Acad Sci 119(6): e2112663119.
- Gutiérrez S, Michalakis Y, Blanc S (2012) Virus population bottlenecks during within-host progression and host-to-host transmission. Curr Opin Virol 2(5): 546-555.
© 2025, Efe Sezgin. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






