- Submissions

Full Text
Cohesive Journal of Microbiology & Infectious Disease
Microbiome Profiling for Chronic Disease: Redefining the Future of Diagnosis-An Update
Swarup K Chakrabarti1* and Dhrubajyoti Chattopadhyay1,2
1H P Ghosh Research Center, India
2Sister Nivedita University, India
*Corresponding author: Dr Swarup K Chakrabarti, PhD, H P Ghosh Research Center, HIDCO (II), EK Tower, New Town, Kolkata, West Bengal 700161, India
Submission: August 26, 2025;Published: September 26, 2025

ISSN 2578-0190 Volume7 issues 5
Abstract
The human microbiome is emerging as a central player in chronic disease management. Breakthroughs in microbiome profiling-particularly spatial microbiomics and Artificial Intelligence (AI)-driven analytics-are reshaping how diseases are diagnosed, monitored and treated. This review examines the transformative potential of microbiome profiling in managing conditions such as type 2 diabetes, cardiovascular disease, autoimmune disorders and neurodegenerative diseases. It highlights the convergence of spatial diagnostics, AI and microbiome-based biomarkers as the foundation of next-generation precision medicine. High-resolution microbiome mapping enables earlier detection, more accurate diagnostics and tailored therapeutic strategies. AI algorithms unravel complex microbiome datasets, revealing diseaserelevant patterns, while microbial biomarkers provide actionable therapeutic targets. Case studies in Crohn’s disease and colorectal cancer illustrate how microbiome profiling can inform personalized interventions, including probiotics and dietary modulation. Collectively, these innovations are driving a shift toward precision healthcare grounded in individual microbial architectures-promising improved patient outcomes and heralding a new era of personalized care.
Keywords: Microbiome profiling; Chronic disease management; Spatial microbiomics; Precision medicine; Microbial biomarkers; Next-generation sequencing metagenomics
Introduction
The human microbiome-comprising trillions of bacteria, archaea, fungi and virusesfunctions as a dynamic, adaptive interface between the host and its environment [1-3]. Beyond its well-established role in digestion, this vast microbial ecosystem contributes to immune modulation, metabolic homeostasis and neuroendocrine communication, shaping health trajectories from early development through late life [4-6]. Advances in high-resolution Next- Generation Sequencing (NGS) and spatial microbiomics reveal that dysbiosis-an imbalance in microbial composition-may not merely result from chronic disease but can act as an upstream driver, altering immune tone, metabolic flux and epithelial barrier integrity long before clinical disease onset [7-11].
Large-scale initiatives such as the Human Microbiome Project (HMP) and the International Human Microbiome Consortium (IHMC) have generated critical insights into disease-associated microbial signatures across diverse body sites [12]. However, emerging evidence indicates that understanding where microbes reside (spatial niches) and how they function (functional states) is as important as identifying which microbes are present [13,14]. Increasingly, the microbiome is conceptualized as an integrated, multi-organ network node, linked by bidirectional “axes” connecting the gut with the brain, lung and heart, as well as with the skin, musculoskeletal system, endocrine glands and even the tumor microenvironment [15-20].
Technological advances, including Polymerase Chain Reaction (PCR) and 16S rRNA sequencing, have revolutionized microbial analysis and accelerated the pace of microbiome research [4,21]. Currently, more than 20,000 publicly available metagenomic datasets and over 130,000 complete bacterial genomes constitute an unprecedented resource, underscoring the need for integrative approaches to elucidate microbiome-mediated regulation of chronic disease [22,23]. Artificial intelligence (AI), particularly Machine Learning (ML), is advancing microbiome profiling by identifying microbial biomarkers that enhance disease prediction and support personalized treatment strategies [24,25]. Spatial diagnostic platforms-such as spatial transcriptomics and spatial microbiomics-enable in situ mapping of microbes within their native tissue environments, revealing microbe–host cell interactions and informing precision medicine [26-28].
These conceptual and technological advances are converging: third-generation sequencing [29,30], AI-driven biomarker discovery [24,25] and spatial diagnostics [26-28] now allow tissuelevel mapping of microbial-host interactions, real-time profiling in clinical contexts and prediction of therapeutic responses. This integration lays the foundation for microbiome-informed precision medicine, in which diagnosis, prognosis and therapy are guided by an individual’s dynamic microbial architecture-monitored not as a single snapshot, but longitudinally. The present review examines these advances, emphasizing how emerging technologies, AI and biomarker-driven interventions can translate microbiome science into proactive chronic disease management.
Technological Advances in Microbiome Profiling
Distinguishing the role of the microbiome in diseases requires microbiome profiling from different sites in the human body. In such cases, the gut microbiome is typically assessed through stool samples; oral samples from the microbiome can be extracted from saliva or with dental swabs; skin samples from the skin microbiome can be taken from skin swabs; and the respiratory microbiome samples can be collected using nasal or throat swabs [31-33]. Researchers transfer these samples under sterile conditions, preserve them properly and analyze them using molecular techniques to ensure reliability. Researchers use advanced sequencing methods, like 16S rRNA and shotgun metagenomic sequencing, to analyze microbial DNA or RNA and study its link to disease.
Traditional culturing techniques are increasingly being replaced by molecular methods. For example, 16S rRNA gene sequencing identifies and quantifies bacterial species by targeting specific regions of the 16S gene [3-5]. In contrast, shotgun metagenomic sequencing analyzes the entire microbial DNA, enabling the detection of a wider range of microorganisms, including those previously unculturable [34]. Researchers primarily use Next- Generation Sequencing (NGS) to profile the microbiome by analyzing hypervariable sub-regions of the 16S rRNA gene [35]. This high-throughput sequencing technique amplifies specific gene regions, allowing simultaneous sequencing of short DNA fragments (150-250 base pairs). Researchers typically target the V4 or V3-V4 hypervariable regions to classify bacteria at the genus level. However, this method has limitations, including unreliable species-level classification due to sequence homology, biases from sequencing platforms and inconsistencies in data processing across different software [36,37]. Lastly, sequencing of the very small, highly variable parts among these total 1542 nucleotides of the 16S rRNA gene precludes identification of important genetic variation necessary to separate closely related bacterial species [38].
Figure 1: Microbiome profiling workflow. The flowchart outlines the process of microbiome profiling, beginning with sample collection from the gut, oral cavity, skin, and respiratory sites. Samples are collected using sterile techniques, preserved and preferably maintained at low temperature, depending on the sample type, prior to molecular analyses. Sequencing approaches such as 16S rRNA gene sequencing and shotgun metagenomics are employed to identify microbial communities within samples. Methodological advances, including full-length 16S rRNA sequencing, Whole-Genome Sequencing (WGS) and nanopore sequencing, have further enhanced the resolution and accuracy of microbiome profiling. Ultimately, spatial diagnostic approaches provide deeper insights into host-microbiome interactions.
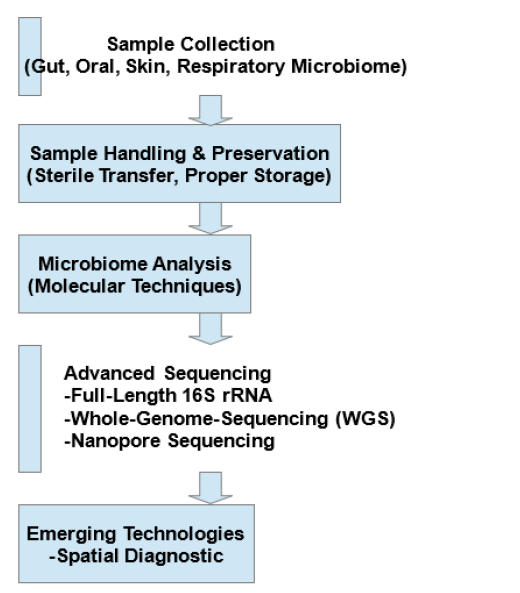
To overcome these challenges, researchers are refining fulllength 16S rRNA gene sequencing and Whole-Genome Sequencing (WGS) with shotgun metagenomics [39]. These methods became feasible with the advent of the third-generation sequencing technologies that allow for more accurate characterization of microbes by capturing the complete genetic landscape of microbial communities. The improved sequencing technologies, especially nanopore sequencing, have changed the very nature of microbiome profiling by enabling real-time and portable sequencing fit for almost point-of-care diagnostics [40]. In addition, emerging spatial diagnostics allow the intratissue investigation of microbial communities [41]. Figure 1 describes the microbiome profiling workflow, outlining steps from sample collection to advanced sequencing and spatial diagnostics for studying host-microbiome interaction.
Spatial diagnostics for chronic disease management
Spatial transcriptomics and microbiomics track gene expression and microbial activity in tissues, revealing how microbes interact with host cells [26,41,42]. This spatial perspective sheds light on the contribution of these microbial communities to disease progression, enhancing accurate diagnostics and tailored treatment. Microorganisms rarely exist alone; they form complex communities that interact with each other and their hosts. These interactions can be cooperative (mutualistic or symbiotic) or antagonistic (competitive or inhibitory), both of which help maintain ecosystem stability in plants, animals and humans [43,44]. The spatial arrangement of microbial communities plays a crucial role in their ability to colonize a host and influence health or disease. Studies show that host-associated microbes form distinct patterns, with some species clustering together while others remain separate [45,46]. This arrangement influences competition among microbes, sharing resources and interaction with the immune system of the host. This makes it all the more important to understand the complex host-microbiome-microbiome interactome: the web of interactions among microorganisms in the host and their collective effects on the physiology of the host [47,48].
The spatial arrangement of microbes is important for diagnosing and managing chronic diseases like Inflammatory Bowel Disease (IBD), T2D, neurodegenerative diseases (Alzheimer’s and Parkinson’s) and autoimmune disorders [49- 51]. Unlike acute infections caused by a single pathogen, these chronic diseases result from microbial imbalances (dysbiosis). Understanding microbial spatial patterns can help identify diseaserelated changes and guide targeted treatments. For example, in IBD, pathogenic bacteria can frequently form biofilm in inflamed areas of the gut, while beneficial microbes may be diminished or displaced [52,53]. Therefore, spatial microbiomics could alert us to some of these changes earlier, allowing for more precise detection and classification of disease. In Colorectal Cancer (CRC), certain microbes selectively colonize tumor sites. Using imaging and spatial transcriptomics, scientists can map these microbes to improve early detection and risk assessment [54,55]. Such knowledge can generate strategies for personalized treatment beyond diagnosis. If specific microbial interactions cause disease, spatial diagnosis can help develop targeted probiotics or microbiome-based therapies [56,57]. For example, in metabolic disorders, bringing back the beneficial microbial communities into their correct spatial niches would rebalance the host metabolism. Moreover, it would improve drug delivery, allowing probiotics or small-molecule therapies to target disease sites more effectively with fewer side effects [58,59]. Another important issue is the monitoring of microbial events over time to assess disease progression, especially in chronic diseases that develop over years or even decades. Mapping host-microbiome interactions at the cellular level can provide insight into how microbial dysbiosis triggers or exacerbates immune defects, thus opening our understanding to new and promising routes for therapies in inflammatory and autoimmune diseases [60,61].
Emerging technologies like in situ sequencing, single-cell RNA sequencing, and spatial transcriptomics are rapidly advancing toward clinical use in spatial microbiomics. More specifically, at the level of resolution attained with these modalities, we have discovered a new and powerful means to determine diseases, track them over time and treat non-infectious chronic diseases by high-resolution maps of microbial communities present within tissues. These communities exist not only within the host but also in a structured manner when recognized. This could lead to sophisticated strategies for disease prevention and management, heralding a new age of ‘microbiome-informed’ precision medicine [62-64].
In summary, advanced microbiome profiling, like 16S rRNA sequencing and spatial microbiomics, is transforming disease diagnosis and treatment. This technology champions high-fidelity microbial characterization and real-time diagnosis of chronic disease. Spatial diagnostics then personalize therapies towards better, more effective, targeted and precision medicine. The future of microbiome science is indeed bright, with breakthroughs paving the way for tailored, microbially based treatments that could revolutionize clinical practice and improve patients’ lives.
Integration of Artificial Intelligence in Microbiome Analysis
It has been very challenging to understand a vast amount of biological data concerned with microorganisms and their prevalence. But due to advancements in massive sequencing, ML, which is a subsection of AI, can be applied to process enormous amounts of data concerning microbes and arrive at diagnoses for a range of diseases [24,25]. Extracting useful information from vast and complex microbiome data requires powerful computing resources. In such scenarios, AI and ML, in particular, have proven essential. Large data sets of the microbiome may be examined through AI algorithms that can identify associations that standard statistical techniques may be unable to identify [65,66]. AI can provide a deep understanding of the role of the microbiome in health and disease by integrating data from genomes, transcriptomics and metabolomics. For identification of microbial biomarkers associated with chronic disease, the microbiome data can be leveraged to perform training of ML algorithms, for instance, random forest, Support Vector Machine (SVM), and clustering algorithms [67,68]. AI-driven methods use microbial profiles to uncover hidden disease subgroups, improving diagnosis and enabling personalized treatments. By integrating clinical data with microbiome data, AI can predict disease trajectories. Such AI algorithms, for instance, can also track the change in microbiome composition with respect to time to predict the future course of diseases such as T2D [69,70]. AI can also predict which patients will be most likely to benefit from microbiome-based therapies, such as Fecal Microbiota Transplantation (FMT) or probiotics, thus providing more customized and effective medical care [71,72].
Figure 1: Artificial Intelligence (AI) and Machine Learning (ML) in Microbiome Research: From Data Processing to Disease Prediction. This flowchart illustrates the role of AI and ML in microbiome research, spanning data processing to disease prediction. AI algorithms are applied to large-scale microbiome datasets to uncover previously unrecognized associations between microbial patterns and disease states. ML models, including Random Forest (RF) and Support Vector Machine (SVM), are utilized to identify microbial biomarkers and predict disease trajectories, thereby supporting the development of personalized therapeutic strategies. Convolutional Neural Networks (CNNs) further enhance pattern recognition within high-dimensional sequencing data, improving diagnostic accuracy, risk stratification and targeted interventions. By integrating microbiome profiles with clinical data, AI-driven approaches advance precision medicine and facilitate microbiome-based therapeutic applications.
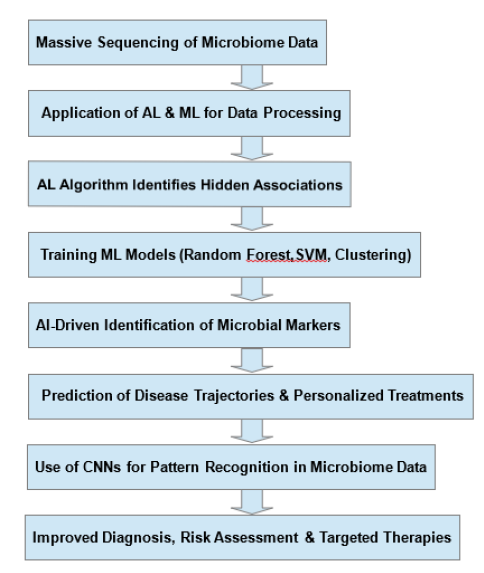
Moreover, Convolutional Neural Networks (CNNs) are highly useful for analyzing complex microbiome data, especially highdimensional sequencing reads [73,74]. CNNs learn to identify unique patterns that distinguish microbial populations from one another so that they can be classified into distinct groups such as healthy and ill. CNNs would probably improve the discovery of microbial signatures that are associated with particular diseases because they learn the hierarchical relationships between taxa. In metagenomics, CNNs analyze large sequencing data to link microbial patterns with clinical outcomes, improving disease risk assessment [75,76]. Automated selection of key microbes and identification of disease-related traits will enhance diagnostic and treatment accuracy. CNNs and ML together help study microbes and their effects on health [77,78]. Figure 2 illustrates, in a flowchart format, the role of AI and ML in microbiome research, from data processing to disease prediction.
Microbiome-Based Biomarkers in Chronic Disease Management
The human microbiome interacts with neurobiology, immunity and metabolism, playing a crucial role in both health and disease [79,80]. Microbiome-derived biomarkers offer insights into hostmicrobe interactions and help in diagnosing, predicting and treating diseases [81,82]. Taxa-specific biomarkers highlight microbial shifts in diseases, such as more Fusobacterium nucleatum in colorectal cancer, an altered Firmicutes/Bacteroidetes ratio in obesity, and less Faecalibacterium prausnitzii in IBD [83,84]. Functional biomarkers link microbial genes and metabolism to diseases, such as reduced butyrate synthesis weakening the gut barrier in IBD, TMAO-producing bacteria contributing to heart disease and tryptophan imbalances affecting neurodegenerative conditions [85,86]. Metabolite biomarkers, which include SCFAs, TMAO, and tryptophan derivatives, are responsible for both immunomodulatory effects and the regulation of metabolic homeostasis and neurotransmission [87,88]. Immune-related biomarkers, such as cytokine changes and microbial patterns, show immune disruptions in Inflammatory Bowel Disease (IBD), obesity and autoimmune disorders [89,90]. Microbiota profiling combined with immune cell analysis helps reveal disease mechanisms, improving diagnosis and treatment [90,91].
Microbiome Profiling in Chronic Disease Management: Case Studies
The human microbiome contributes to general health and is increasingly recognized as a contributor to the pathogenesis of a multitude of chronic diseases [7-11]. Recent advances in microbiome profiling reveal microbiota in chronic disease patients and enable targeted interventions [4-6,23-28]. This section explores microbiome profiling as a diagnostic and therapeutic approach, using Crohn’s Disease (CD) and CRC as specific case studies due to the focused nature of this article. CD, a type of IBD, is linked to microbial imbalances that may trigger its onset and severity [92-94]. Research has identified Mycobacteria spp. and various viral infections as key factors in CD development and progression [95,96]. Additionally, CD patients have a disrupted gut microbiome, with higher levels of Bacteroidetes and Escherichia coli but lower levels of Firmicutes spp. and Faecalibacterium prausnitzii, a crucial anti-inflammatory bacterium [97,98]. One of the primary traits of the CD-associated microbiota is the formation of biofilms on the intestinal epithelium [99]. The bacteria growing in individuals diagnosed with CD and Ulcerative Colitis (UC) invade intestinal epithelial cells and stimulate the body’s defense mechanism to produce pro-inflammatory cytokines. Notably, Adherent-Invasive Escherichia Coli (AIEC) is a well-recognized cause of the disease that starts and perpetuates the disease [100,101]. Understanding the microbial compositions of CD facilitates the identification of the ways that inform therapeutic agents such as probiotics, prebiotics and synbiotics, which may be given to patients in order to restore eubiosis and reduce inflammation.
CRC is the third most common cancer in the world today, with an estimated 1.8 million new cases diagnosed in 2018 [102]. Recent microbiome studies have found specific bacterial species that are responsible for the development of CRC. Bacteria linked to CRC include Fusobacterium nucleatum, Enterococcus faecalis, Bacteroides fragilis, Streptococcus gallolyticus and Porphyromonas [103]. It is possible that these bacteria are involved in tumorigenesis through actions such as inducing inflammation, hindering the gut barrier or producing carcinogenic metabolites. Microbiome profiling can help detect microbial biomarkers for CRC, leading to better screening and early detection. This may enable interventions like dietary changes and probiotic therapy [104,105].
Hence, the application of microbiome profiling in the management of chronic diseases has significant promise for personalized medicine strategies. Through the identification of the community structures of the microorganisms unique to each disease stage, healthcare professionals can design targeted interventions that will help the reestablishment of the microbial balance. For CD, possible treatments include probiotic strains, dietary fiber to support their growth or combined symbiotic therapies [106,107]. In CRC, microbiome profiling can guide dietary interventions and predict microbial imbalances linked to cancer risk. Using microbiome profiling throughout disease progression can improve chronic disease management by enabling more accurate diagnosis and personalized treatments [108,109].
Challenges & Limitations
Microbiome profiling has advanced a lot over the years, yet a collection of both technical and biological challenges still remains. Factors such as individual variation, exposome, dietary habits and antibiotic usage are the keys that lead to the complexity of microbiome analyses [110,111]. They add another layer to the challenge, as the microbiome is a dynamic entity that changes over time because of internal and external factors. Hence, linking disease development to microbial profiles requires regular or periodic sample collection, not just a single test. Moreover, contamination of samples, together with the use of inconsistent methodologies and the lack of uniform protocols across all clinical laboratories, make it very challenging to ensure reproducibility and the homogeneity of the results. These factors make it difficult to establish a causal link between microbial communities and health outcomes [112]. It is essential to optimize methods while considering these variables to support the field and obtain more accurate, useful information from microbiome studies.
Furthermore, the microbiome is highly diverse, so it complicates interpreting data obtained from microbiome profiling studies. This occurs because of the collection of techniques, the methods of data analysis, and the reference databases, which are not constant across all analyses [113,114]. As a result, the findings are not uniform. One way of improving the reliability and reproducibility of microbiome research, especially in clinical settings, is by optimizing the protocols for sample collection, processing and data analysis. Also, due to the limited resources of a budget in a clinical setting, this specific article limits its focus to only sequencing-based metagenomics studies. However, this approach fails to account for intricate gene pathways, proteins, and metabolic processes that may drive disease. In the future, the integration of multi-omics approaches, for instance, gene transcript analysis, protein products, and small molecules of microbial or host metabolism, will provide a more comprehensive insight into the microbiome [115,116]. This comprehensive analysis will aid in identifying etiopathogenetic connections of microbiota and host chronic disease conditions, which will allow the microbiome’s functional dynamics and its potential health/disease effects to be better exemplified [117].
Microbiota data in clinical settings raises critical concerns about privacy, consent and data sharing [118]. Given its potential to reveal valuable health and lifestyle insights, strict guidelines for data storage and use are essential. The risk of privacy breaches from microbial profiles may necessitate new genetic privacy laws, imposing stricter scrutiny on microbiome profiling studies. Furthermore, limited access to microbiome diagnostics and therapeutics remains a major barrier to effectively diagnosing and treating healthcare disparities [119]. Microbiota biobanks also have to overcome ethical dilemmas such as sample ownership, the use of specimens for further scientific purposes, and the sharing of future benefits [120,121]. Participants achieve this by fully disclosing their data and knowing who will use it and how. Also, stringent laws that secure the privacy of the participants and advance microbiome research will be ethically sound. By ensuring both the privacy of individuals and the transparency of microbial profiling, we can establish an ethical, inclusive, and forward-thinking approach that benefits all of humanity. Finally, the challenges in applying AI/ML to microbiome data arise from limitations in study design (e.g., causality, population selection, eligibility criteria, and host-related confounders) and from technical variability (e.g., sample collection, storage, DNA extraction, sequencing and data processing). Such factors can introduce bias and ultimately hinder clinical translation [122].
Significance of the Study
This review integrates advances in third-generation sequencing, spatial microbiomics, AI-driven analytics, and biomarker-based therapeutics into a unified framework for microbiome-guided precision medicine. By moving beyond taxonomic cataloguing to emphasize spatially resolved, functionally relevant host-microbemicrobe interactions, it demonstrates how emerging technologies can shift chronic disease care from reactive symptom management to proactive ecological stewardship of health. Grounded in realworld case studies and anchored by the forward-looking concept of “microbiome passports,” it bridges foundational microbiome science with clinical practice to outline a pathway for reproducible, ethical and equitable implementation. This synthesis not only consolidates the current state of the field but also charts a transformative trajectory toward anticipatory, personalized interventions.
Conclusion
Microbiome research is entering a phase where the focus is shifting from merely cataloguing associations to developing actionable, mechanistically informed interventions. The convergence of next-generation microbiome diagnostics, spatial microbiomics, AI-driven analytics, and biomarker-based therapeutics is enabling a new class of precision healthcare tools capable of assessing disease risk, tracking progression, and guiding personalized interventions. In this emerging paradigm, microbial signatures will not only stratify patients by disease subtype but also predict therapeutic responsiveness, allowing optimization of treatment strategies before clinical symptoms manifest. The next frontier lies in developing dynamic, contextspecific microbiome management strategies. This entails longitudinal spatial profiling to track microbial evolution within defined body niches over time, coupled with real-time monitoring via ingestible or wearable biosensors that detect microbiomederived metabolites. It also includes engineering patient-specific microbial consortia to restore or enhance functional capacity and integrating microbiome-aware clinical decision-support systems that combine genomic, exosmic and lifestyle data to generate actionable insights. However, ethical and infrastructural challenges persist-particularly in standardization, equitable access and data privacy-but are addressable through coordinated, interdisciplinary frameworks. Over the next decade, “microbiome passports” could become as routine as blood typing, guiding not only disease management but also drug dosing, dietary planning and preventive health interventions. Finally, integrating microbial ecology into conventional medical practice has the potential to shift chronic disease care from reactive to anticipatory-enabling interventions at the earliest, most reversible stages and establishing microbial stewardship as a cornerstone of human health.
Conflict of Interest
The authors do have anything to declare.
Funding
This research is supported by Bandhan, Kolkata, India.
Author Contribution
Conceptualization: S. K. C.; Formal analysis: S. K. C.; Original draft preparation: S. K. C.; Writing-review and editing: S. K. C. and D. C.; Supervision: S. K. C.; Project administration: S. K. C.; Funding acquisition: S. K. C.
References
- Thursby E, Juge N (2017) Introduction to the human gut microbiota. Biochem J 474(11): 1823-1836.
- Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, et al. (2015) Role of the normal gut microbiota. World J Gastroenterol 21(29): 8787-8803.
- Afzaal M, Saeed F, Shah YA, Hussain M, Rabail R, et al. (2022) Human gut microbiota in health and disease: Unveiling the relationship. Front Microbiol 13: 999001.
- Satam H, Joshi K, Mangrolia U, Waghoo S, Zaidi G, et al. (2024) Next-generation sequencing technology: Current trends and advancements. Biology 12(7): 997.
- Do T, Deng D, Dame-Teixeira N (2024) Editorial: Applications of Next Generation Sequencing (NGS) technologies to decipher the oral microbiome in systemic health and disease, volume II. Front Cell Infect Microbiol 14: 1532762.
- Hemmati MA, Monemi M, Asli S, Mohammadi S, Foroozanmehr B, et al. (2024) Using new technologies to analyze gut microbiota and predict cancer risk. Cells 13(23): 1987.
- Carding S, Verbeke K, Vipond DT, Corfe BM, Owen LJ (2015) Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis 26: 26191.
- Zaky A, Glastras SJ, Wong MY, Pollock CA, Saad S (2021) The role of the gut microbiome in diabetes and obesity-related kidney disease. Int J Mol Sci 22(17): 9641.
- Chang SH, Choi Y (2023) Gut dysbiosis in autoimmune diseases: Association with mortality. Front Cell Infect Microbiol 13: 1157918.
- Singh R, Zogg H, Wei L, Bartlett A, Ghoshal UC, et al. (2021) Gut microbial dysbiosis in the pathogenesis of gastrointestinal dysmotility and metabolic disorders. J Neurogastroenterol Motil 27(1): 19-34.
- Toor D, Wsson MK, Kumar P, Karthikeyan G, Kaushik NK, et al. (2019) Dysbiosis disrupts gut immune homeostasis and promotes gastric diseases. Int J Mol Sci 20(10): 2432.
- Peterson J, Garges S, Giovanni M, McInnes P, Wang L, et al. (2009) The NIH human microbiome project. Genome Res 19(12): 2317-2323.
- Aggarwal N, Kitano S, Puah GR, Kittelmann S, Hwang IY, et al. (2023) Microbiome and human health: Current understanding, engineering and enabling technologies. Chem Rev 123(1): 31-72.
- Kamel M, Aleya S, Alsubih M, Aleya L (2024) Microbiome dynamics: A paradigm shift in combatting infectious diseases. J Pers Med 14(2): 217.
- Zhu X, Han Y, Du J, Liu R, Jin K, et al. (2017) Microbiota-gut-brain axis and the central nervous system. Oncotarget 8(32): 53829-53838.
- Ashique S, Mohanto S, Ahmed MG, Mishra N, Garg A, et al. (2024) Gut-brain axis: A cutting-edge approach to target neurological disorders and potential synbiotic application. Heliyon 10(13): e34092.
- Ziaka M, Exadaktylos A (2024) Exploring the lung-gut direction of the gut-lung axis in patients with ARDS. Crit Care 28(1): 179.
- Druszczynska M, Sadowska B, Kulesza J, Gąsienica-Gliwa N, Kulesza E, et al. (2024) The intriguing connection between the gut and lung microbiomes. Pathogens 13(11): 1005.
- Sagmeister A, Matter CM, Stähli BE, Scharl M (2024) The gut-heart axis: Effects of intestinal microbiome modulation on cardiovascular disease-ready for therapeutic interventions? Int J Mol Sci 25(24): 13529.
- Zhu T, Goodarzi MO (2020) Metabolites linking the gut microbiome with risk for type 2 diabetes. Curr Nutr Rep 9(2): 83-93.
- Jansen GJ, Schouten GP, Wiersma M (2024) Advancements in analytical methods for studying the human gut microbiome. J Biol Methods 12(1): e99010038.
- Pasolli E, Asnicar F, Manara S, Zolfo M, Karcher N, et al. (2019) Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography and lifestyle. Cell 176(3): 649-662.
- Zhu J, Tian L, Chen P, Han M, Song L, et al. (2022) Over 50,000 metagenomically assembled draft genomes for the human oral microbiome reveal new taxa. Genomics Proteomics Bioinformatics 20(2): 246-259.
- Papoutsoglou G, Tarazona S, Lopes MB, Klammsteiner T, Ibrahimi E, et al. (2023) Machine learning approaches in microbiome research: Challenges and best practices. Front Microbiol 14: 1261889.
- Malwe AS, Sharma VK (2023) Application of artificial intelligence approaches to predict the metabolism of xenobiotic molecules by human gut microbiome. Front Microbiol 14: 1254073.
- Saarenpää S, Shalev O, Ashkenazy H, Carlos V, Lundberg DS, et al. (2024) Spatial metatranscriptomics resolves host-bacteria-fungi interactomes. Nat Biotechnol 42(9): 1384-1393.
- Di Marco F, Nicola F, Giannese F, Saliu F, Tonon G, et al. (2024) Dual spatial host-bacterial gene expression in Mycobacterium abscessus respiratory infections. Commun Biol 7(1): 1287.
- Adade EE, Al Lakhen, Lemus AA, Valm AM (2021) Recent progress in analyzing the spatial structure of the human microbiome: Distinguishing biogeography and architecture in the oral and gut communities. Curr Opin Endocr Metab Res 18: 275-283.
- Purushothaman S, Meola M, Egli A (2022) Combination of whole genome sequencing and metagenomics for microbiological diagnostics. Int J Mol Sci 23(17): 9834.
- Chen P, Sun Z, Wang J, Liu X, Bai Y, et al. (2023) Portable nanopore-sequencing technology: Trends in development and applications. Front Microbiol 14: 1043967.
- Kodukula K, Faller DV, Harpp DN, Kanara I, Pernokas J, et al. (2017) Gut Microbiota and salivary diagnostics: The mouth is salivating to tell us something. Biores Open Access 6(1): 123-132.
- Schmartz GP, Rehner J, Gund MP, Keller V, Molano LG, et al. (2024) Decoding the diagnostic and therapeutic potential of microbiota using pan-body pan-disease microbiomics. Nat Commun 15(1): 8261.
- Furusawa C, Tanabe K, Ishii C, Kagata N, Tomita M, et al. (2021) Decoding gut microbiota by imaging analysis of fecal samples. iScience 24(12): 103481.
- Björnberg A, Nestor D, Peker N, Sinha B, Couto N, et al. (2025) Critical steps in shotgun metagenomics-based diagnosis of bloodstream infections using nanopore sequencing. APMIS 133(1): e13511.
- López-Aladid R, Fernández-Barat L, Alcaraz-Serrano V, Bueno-Freire L, Vázquez N, et al. (2023) Determining the most accurate 16S rRNA hypervariable region for taxonomic identification from respiratory samples. Sci Rep 13(1): 3974.
- Zhang H, Wang X, Chen A, Li S, Tao R, et al. (2024) Comparison of the full-length sequence and sub-regions of 16S rRNA gene for skin microbiome profiling. mSystems 9(7): e0039924.
- Leontidou K, Abad-Recio IL, Rubel V, Filker S, Däumer M, et al. (2023) Simultaneous analysis of seven 16S rRNA hypervariable gene regions increases efficiency in marine bacterial diversity detection. Environ Microbiol 25(12): 3484-3501.
- Church DL, Cerutti L, Gürtler A, Griener T, Zelazny A, et al. (2020) Performance and application of 16S rRNA gene cycle sequencing for routine identification of bacteria in the clinical microbiology laboratory. Clin Microbiol Rev 33(4): e00053-19.
- Purushothaman S, Meola M, Egli A (2022) Combination of whole genome sequencing and metagenomics for microbiological diagnostics. Int J Mol Sci 23(17): 9834.
- Chen P, Sun Z, Wang J, Liu X, Bai Y, et al. (2023) Portable nanopore-sequencing technology: Trends in development and applications. Front Microbiol 14: 1043967.
- Adade EE, Al Lakhen, Lemus AA, Valm AM (2021) Recent progress in analyzing the spatial structure of the human microbiome: Distinguishing biogeography and architecture in the oral and gut communities. Curr Opin Endocr Metab Res 18: 275-283.
- Di Marco F, Nicola F, Giannese F, Saliu F, Tonon G, et al. (2024) Dual spatial host-bacterial gene expression in Mycobacterium abscessus respiratory infections. Commun Biol 7(1): 1287.
- Weiland-Bräuer N (2021) Friends or foes-microbial interactions in nature. Biology 10(6): 496.
- Moënne-Loccoz Y, Mavingui P, Combes C, Normand P, Steinberg C (2014) Microorganisms and biotic interactions. Environmental Microbiology: Fundamentals and Applications 29: 395-444.
- Ruuskanen MO, Sommeria-Klein G, Havulinna AS, Niiranen TJ, Lahti L (2021) Modeling spatial patterns in host-associated microbial communities. Environ Microbiol 23(5): 2374-2388.
- Duncan K, Carey-Ewend K, Vaishnava S (2021) Spatial analysis of gut microbiome reveals a distinct ecological niche associated with the mucus layer. Gut Microbes 13(1): 1874815.
- Collado MC, Cernada M, Baüerl C, Vento M, Pérez-Martínez G (2012) Microbial ecology and host-microbiota interactions during early life stages. Gut Microbes 3(4): 352-365.
- Zheng D, Liwinski T, Elinav E (2020) Interaction between microbiota and immunity in health and disease. Cell Res 30(6): 492-506.
- Kim JS, Chen MH, Wang HE, Lu CL, Wang YP, et al. (2023) Inflammatory bowel disease and neurodegenerative diseases. Gut Liver 17(4): 495-504.
- Günther C, Rothhammer V, Karow M, Neurath M, Winner B (2021) The gut-brain axis in inflammatory bowel disease-current and future perspectives. Int J Mol Sci 22(16): 8870.
- Duda-Madej A, Stecko J, Szymańska N, Miętkiewicz A, Szandruk-Bender M (2024) Amyloid, crohn’s disease and alzheimer’s disease-are they linked? Front Cell Infect Microbiol 14: 1393809.
- Palandurkar GS, Kumar S (2023) Biofilm's impact on inflammatory bowel diseases. Cureus 15(9): e45510.
- Vidal-Gallardo A, Benítez JE, Rios L, Meza LF, Pérez RA, et al. (2024) The role of gut microbiome in the pathogenesis and the treatment of inflammatory bowel diseases. Cureus 16(2): e54569.
- Carranza FG, Diaz FC, Ninova M, Velazquez-Villarreal E (2024) Current state and future prospects of spatial biology in colorectal cancer. Front Oncol 14: 1513821.
- Long J, Wang J, Xiao C, You F, Jiang Y, et al. (2024) Intratumoral microbiota in colorectal cancer: Focus on specific distribution and potential mechanisms. Cell Commun Signal 22(1): 455.
- Hitch TC, Hall LJ, Walsh SK, Leventhal GE, Slack E, et al. (2022) Microbiome-based interventions to modulate gut ecology and the immune system. Mucosal Immunol 15(6): 1095-1113.
- Robinson CM, Short NE, Riglar DT (2022) Achieving spatially precise diagnosis and therapy in the mammalian gut using synthetic microbial gene circuits. Front Bioeng Biotechnol 10: 959441.
- Chen A, Gong Y, Wu S, Du Y, Liu Z, et al. (2025) Navigating a challenging path: Precision disease treatment with tailored oral nano-armor-probiotics. J Nanobiotechnology 23(1): 72.
- Li C, Wang ZX, Xiao H, Wu FG (2024) Intestinal delivery of probiotics: Materials, strategies and applications. Adv Mater 36(32): e2310174.
- Ahern PP, Maloy KJ (2020) Understanding immune-microbiota interactions in the intestine. Immunology 159(1): 4-14.
- Ma Z, Zuo T, Frey N, Rangrez AY (2024) A systematic framework for understanding the microbiome in human health and disease: From basic principles to clinical translation. Signal Transduct Target Ther 9(1): 237.
- Kamel M, Aleya S, Alsubih M, Aleya L (2024) Microbiome dynamics: A paradigm shift in combatting infectious diseases. J Pers Med 14(2): 217.
- Mendez KM, Reinke SN, Kelly RS, Chen Q, Su M, et al. (2025) A roadmap to precision medicine through post-genomic electronic medical records. Nat Commun 16(1): 1700.
- Jangi S, Hecht G (2024) Microbiome 2.0: Lessons from the 2024 gut microbiota for health world summit. Gut Microbes 16(1): 2400579.
- Probul N, Huang Z, Saak CC, Baumbach J, List M (2024) AI in microbiome-related healthcare. Microb Biotechnol 17(11): e70027.
- Moreno-Indias I, Lahti L, Nedyalkova M, Elbere I, Roshchupkin G, et al. (2021) Statistical and machine learning techniques in human microbiome studies: Contemporary challenges and solutions. Front Microbiol 12: 635781.
- Sun T, Niu X, He Q, Chen F, Qi RQ (2023) Artificial intelligence in microbiomes analysis: A review of applications in dermatology. Front Microbiol 14: 1112010.
- Kumar B, Lorusso E, Fosso B, Pesole G (2024) A comprehensive overview of microbiome data in the light of machine learning applications: Categorization, accessibility and future directions. Front Microbiol 15: 1343572.
- Li P, Luo H, Ji B, Nielsen J (2022) Machine learning for data integration in human gut microbiome. Microb Cell Fact 21(1): 241.
- Mohsen F, Al-Absi HR, Yousri NA, El Hajj N, Shah Z (2023) A scoping review of artificial intelligence-based methods for diabetes risk prediction. NPJ Digit Med 6(1): 197.
- McCoubrey LE, Elbadawi M, Orlu M, Gaisford S, Basit AW (2021) Harnessing machine learning for development of microbiome therapeutics. Gut Microbes 13(1): 1-20.
- Schupack DA, Mars RA, Voelker DH, Abeykoon JP, Kashyap PC (2022) The promise of the gut microbiome as part of individualized treatment strategies. Nat Rev Gastroenterol Hepatol 19(1): 7-25.
- Vaz JM, Balaji S (2021) Convolutional Neural Networks (CNNs): Concepts and applications in pharmacogenomics. Mol Divers 25(3): 1569-1584.
- Przymus P, Rykaczewski K, Martín-Segura A, Truu J, Kolev M, et al. (2025) Deep learning in microbiome analysis: A comprehensive review of neural network models. Front Microbiol 15: 1516667.
- Liu Y, Ma Y (2024) Clinical applications of metagenomics next-generation sequencing in infectious diseases. J Zhejiang Univ Sci B 25(6): 471-484.
- Roy G, Prifti E, Belda E, Zucker JD (2024) Deep learning methods in metagenomics: A review. Microb Genom 10(4): 001231.
- Marcos-Zambrano LJ, Karaduzovic-Hadziabdic K, Turukalo TL, Przymus P, Trajkovik V, et al. (2021) Applications of machine learning in human microbiome studies: A review on feature selection, biomarker identification, disease prediction and treatment. Front Microbiol 12: 634511.
- Chen AT, Wu X, Ye G, Li W (2024) Editorial: machine learning and deep learning applications in pathogenic microbiome research. Front Cell Infect Microbiol 14: 1429197.
- Juarez VM, Montalbine AN, Singh A (2022) Microbiome as an immune regulator in health, disease and therapeutics. Adv Drug Deliv Rev 188: 114400.
- Fung TC, Olson CA, Hsiao EY (2017) Interactions between the microbiota, immune and nervous systems in health and disease. Nat Neurosci 20(2): 145-155.
- Hajjo R, Sabbah DA, Al Bawab AQ (2022) Unlocking the potential of the human microbiome for identifying disease diagnostic biomarkers. Diagnostics 12(7): 1742.
- Yavuz BG, Datar S, Chamseddine S, Mohamed YI, Pelusa ML, et al. (2023) The gut microbiome as a biomarker and therapeutic target in hepatocellular carcinoma. Cancers 15(19): 4875.
- Pandey H, Tang DW, Wong SH, Lal D (2023) Gut microbiota in colorectal cancer: Biological role and therapeutic opportunities. Cancers 15(3): 866.
- Li J, Zhang AH, Wu FF, Wang XJ (2022) Alterations in the gut microbiota and their metabolites in colorectal cancer: Recent progress and future prospects. Front Oncol 12: 841552.
- Gao W, Gao X, Zhu L, Gao S, Sun R, et al. (2023) Multimodal metagenomic analysis reveals microbial single nucleotide variants as superior biomarkers for early detection of colorectal cancer. Gut Microbes 15(2): 2245562.
- Zhao Z, Chen J, Zhao D, Chen B, Wang Q, et al. (2024) Microbial biomarker discovery in parkinson's disease through a network-based approach. NPJ Parkinsons Dis 10(1): 203.
- Losol P, Wolska M, Wypych TP, Yao L, O'Mahony L, et al. (2024) A cross talk between microbial metabolites and host immunity: Its relevance for allergic diseases. Clin Transl Allergy 14(2): e12339.
- Lu S, Wang C, Ma J, Wang Y (2024) Metabolic mediators: Microbial-derived metabolites as key regulators of anti-tumor immunity, immunotherapy and chemotherapy. Front Immunol 15: 1456030.
- Mousa WK, Chehadeh F, Husband S (2022) Microbial dysbiosis in the gut drives systemic autoimmune diseases. Front Immunol 13: 906258.
- Liu C, Fu L, Wang Y, Yang W (2024) Influence of the gut microbiota on immune cell interactions and cancer treatment. J Transl Med 22(1): 939.
- Li Z, Xiong W, Liang Z, Wang J, Zeng Z, et al. (2024) Critical role of the gut microbiota in immune responses and cancer immunotherapy. J Hematol Oncol 17(1): 33.
- Duda-Madej A, Stecko J, Szymańska N, Miętkiewicz A, Szandruk-Bender M (2024) Amyloid, crohn's disease and alzheimer's disease-are they linked? Front Cell Infect Microbiol 14: 1393809.
- Palandurkar GS, Kumar S (2023) Biofilm's Impact on Inflammatory Bowel Diseases. Cureus 15(9): e45510.
- Vidal-Gallardo A, Benítez JE, Rios LF, Meza LF, Pérez RA, et al. (2024) The role of gut microbiome in the pathogenesis and the treatment of inflammatory bowel diseases. Cureus 16(2): e54569.
- McNees AL, Markesich D, Zayyani NR, Graham DY (2015) Mycobacterium paratuberculosis as a cause of crohn's disease. Expert Rev Gastroenterol Hepatol 9(12): 1523-1534.
- Naser SA, Sagramsingh SR, Naser AS, Thanigachalam S (2014) Mycobacterium avium subspecies paratuberculosis causes crohn's disease in some inflammatory bowel disease patients. World J Gastroenterol 20(23): 7403-7415.
- Khan I, Ullah N, Zha L, Bai Y, Khan A, et al. (2019) Alteration of gut microbiota in Inflammatory Bowel Disease (IBD): Cause or consequence? IBD treatment targeting the gut microbiome. Pathogens 8(3): 126.
- Ocansey DK, Hang S, Yuan X, Qian H, Zhou M, et al. (2023) The diagnostic and prognostic potential of gut bacteria in inflammatory bowel disease. Gut Microbes 15(1): 2176118.
- Arias SL, Brito IL (2021) Biophysical determinants of biofilm formation in the gut. Curr Opin Biomed Eng 18: 100275.
- Rolhion N, Darfeuille-Michaud A (2007) Adherent-invasive Escherichia coli in inflammatory bowel disease. Inflamm Bowel Dis 13(10): 1277-83.
- Zheng L, Duan SL, Dai YC, Wu SC (2022) Role of adherent invasive Escherichia coli in pathogenesis of inflammatory bowel disease. World J Clin Cases 10(32): 11671-11689.
- (2019) IARC working group on the evaluation of cancer-preventive interventions. Colorectal cancer screening. Lyon (FR): International Agency for Research on Cancer; Colorectal Cancer.
- Rebersek M (2021) Gut microbiome and its role in colorectal cancer. BMC Cancer 21(1): 1325.
- Zwezerijnen-Jiwa FH, Sivov H, Paizs P, Zafeiropoulou K, Kinross J (2023) A systematic review of microbiome-derived biomarkers for early colorectal cancer detection. Neoplasia 36: 100868.
- Chen G, Ren Q, Zhong Z, Li Q, Huang Z, et al. (2024) Exploring the gut microbiome's role in colorectal cancer: Diagnostic and prognostic implications. Front Immunol 15: 1431747.
- Yadav MK, Kumari I, Singh B, Sharma KK, Tiwari SK (2022) Probiotics, prebiotics and synbiotics: Safe options for next-generation therapeutics. Appl Microbiol Biotechnol 106(2): 505-521.
- Simon E, Călinoiu LF, Mitrea L, Vodnar DC (2021) Probiotics, prebiotics and synbiotics: Implications and beneficial effects against irritable bowel syndrome. Nutrients 13(6): 2112.
- Aggarwal N, Kitano S, Puah GR, Kittelmann S, Hwang IY, et al. (2023) Microbiome and human health: Current understanding, engineering and enabling technologies. Chem Rev 123(1): 31-72.
- Pitashny M, Kesten I, Shlon D, Hur DB, Bar-Yoseph H (2025) The future of microbiome therapeutics. Drugs 85(2): 117-125.
- Merra G, Gualtieri P, Placa GL, Frank G, Morte DD, et al. (2024) The relationship between exposome and microbiome. Microorganisms 12(7): 1386.
- Ahn J, Hayes RB (2021) Environmental influences on the human microbiome and implications for noncommunicable disease. Annu Rev Public Health 42: 277-292.
- Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, et al. (2014) Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 12: 87.
- Bokulich NA, Robeson MS (2024) Bioinformatics challenges for profiling the microbiome in cancer: Pitfalls and opportunities. Trends Microbiol 32(12): 1163-1166.
- Tito RY, Verbandt S, Vazquez MA, Lahti L, Verspecht C, et al. (2024) Microbiome confounders and quantitative profiling challenge predicted microbial targets in colorectal cancer development. Nat Med 30(5): 1339-1348.
- Chetty A, Blekhman R (2024) Multi-omic approaches for host-microbiome data integration. Gut Microbes 16(1): 2297860.
- Franzosa EA, Hsu T, Sirota-Madi A, Shafquat A, Abu-Ali G, et al. (2015) Sequencing and beyond: Integrating molecular 'omics' for microbial community profiling. Nat Rev Microbiol 13(6): 360-372.
- Pires L, González-Paramás AM, Heleno SA, Calhelha RC (2024) The role of gut microbiota in the etiopathogenesis of multiple chronic diseases. Antibiotics 13(5): 392.
- Shin A, Xu H (2022) Privacy risks in microbiome research: Public perspectives before and during a global pandemic. Ethics Hum Res 44(4): 2-13.
- Acharjee A, Singh U, Choudhury SP, Gkoutos GV (2022) The diagnostic potential and barriers of microbiome-based therapeutics. Diagnosis 9(4): 411-420.
- Ejtahed HS, Parsa M, Larijani B (2023) Ethical challenges in conducting and the clinical application of human microbiome research. J Med Ethics Hist Med 16: 5.
- Annaratone L, Palma GD, Bonizzi G, Sapino A, Botti G, et al. (2021) Basic principles of biobanking: From biological samples to precision medicine for patients. Virchows Arch. 479(2): 233-246.
- Rozera T, Pasolli E, Segata N, Ianiro G (2025) Machine learning and artificial intelligence in the multi-omics approach to Gut microbiota. Gastroenterology 169(3): 487-501.
© 2025, Swarup K Chakrabarti. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






