- Submissions

Full Text
Cohesive Journal of Microbiology & Infectious Disease
The Evaluation of the Speed-Oligo® Mycobacteria Assay for Identification of Mycobacterium spp. from Smear Positive and Negative Sputum Samples
Gülnur Tarhan1*, İsmail Ceyhan2, Hülya Şimşek3, Özgül Kisa4 and Yusuf Özay5
1 Department of Medical Microbiology, Adıyaman University, Turkey
2 Atatürk Chest Diseases and Thoracic Surgery Training and Research Hospital, Turkey
3 National Public Health Instıtute, Tuberculosis Reference and Research Laboratory, Turkey
4 Department of Medical Microbiology, Gulhane Military Medical Academy, Turkey
5 Department of Medical Biology, Adiyaman University, Turkey
*Corresponding author:Gülnur Tarhan, Faculty of Medicine, Department of Medical Microbiology, Adıyaman University, Turkey
Submission: July 30, 2018; Published: September 24, 2018

ISSN 2578-0190 Volume2 Issue2
Abstract
Speed-oligo® Mycobacteria is an oligochromatographic test for the qualitative detection of Mycobacterium genus and the species of M. tuberculosis complex, directly in clinical samples. It is based on polymerase chain reaction targeting 16S rRNA and 16S-23S rRNA regions and double-reverse hybridization on a dipstick using probes bound to colloidal gold and to the membrane. In this study, we evaluated 25 type reference strains of mycobacteria (18 non-tuberculosis mycobacteria, M. tuberculosis H37Rv and M. tuberculosis H37Ra), 60 sputum samples (40 smear positive, 20 smear negative) collected from patient with suspected TB. All results were compared with microscopy, Löwenstein Jensen culture and Inno-Lipa (GenoType Mycobacterium CM/AS; Hain Lifescience, Germany). All smear positive sputum samples were positive with microscopy, culture and Speed-oligo® Mycobacteria. Of 20 smear negative sputum samples, 7 were culture positive. Of 7 culture positive samples, 3 were positive with microscopy and Speedoligo ® Mycobacteria. It is not effective to identify for M. intermedium, M. kansasii and M. xenopi.
Keywords: Tuberculosis; NTM; Speed oligo; PCR
Introduction
Tuberculosis (TB) is a chronic infectious disease that is common all over the World [1]. In recent years, nontuberculous mycobacteria (NTM) infections are getting increase depend on a immunosuppression, AIDS, malignancy, drug abuse, bad living conditions or medical intervention. NTM causing clinical disease have become increasingly frequent and more varied. The incidence and prevalence of NTM lung disease are increasing worldwide [2-4]. The rapid differentiation between NTM and M. tuberculosis complex (MTBC) is primary importance for effective and accurate treatment protocol [5]. Mycobacteriology laboratory findings have an important role in the diagnosis and control of this disease. MTBC and NTM can not be distinguished by microscopy. Diagnosis with culture method is time consuming. Conventional identification tests at the species level are quite complex and sometimes no definite conclusions can be drawn. PCR based methods are widely used in routine diagnostic laboratories for rapid diagnosis [5-8]. Speed-oligo® Direct Mycobacterium tuberculosis (SO-DMT) is an oligochromatographic test (Vircell SL, Santa Fe, Granada, Spain) for the specific identification of M. tuberculosis complex (MTC) from Mycobacterium genus in respiratory specimens. It is based on polymerase chain reaction targeting 16S rRNA and 16S-23S rRNA regions and double-reverse hybridization on a dipstick using probes bound to colloidal gold and to the membrane. It may represent a fast and easy alternative for differentiating between MTBC and NTM in direct samples at laboratories with standard laboratory equipment (thermocycler and thermoblock) [5-8]. The aim of this study was to evaluate SO-DMT by using 25 type reference strains of mycobacteria (18 non-tuberculosis mycobacteria, M. tuberculosis H37Rv and M. tuberculosis H37Ra), 60 sputum samples (40 smear positive, 20 smear negative) collected from patient with suspected TB.
Material and Methods
Specimen group
In this study, we evaluated 20 type reference strains of mycobacteria (18 non-tuberculosis mycobacteria, M. tuberculosis H37Rv and M. tuberculosis H37Ra), 60 sputum samples (40 smear positive, 20 smear negative) collected from patient with suspected TB.
Specimen processing, microscopy, culture, and storage conditions
Respiratory specimens were decontaminated with 4% NaOHNALC method, directly examined (with Ehrlich-Ziehl-Neelsen staining method), and cultured by Löwentein Jensen medium and Mycobacterium Indicator Growth System (Becton Dickinson,USA) Mycobacteria were identified by the Geno Type Mycobacterium CM/ AS assay (Hain Lifescience, Nehren, Germany). At least one aliquot of all decontaminated specimens was maintained for a maximum of 7 days at 4 °C until assayed with SO-DMT; if the assay procedure was delayed, samples were frozen at -80 °C until their analysis [9- 13].
SO-DMT assay
The SO-DMT assay was carried out as manufactures instructions. Positive and negative controls were used in each test run. The SODMT comprises four steps: DNA extraction, amplification, PCR product detection, and result reading, as detailed below. DNA is extracted from a previously concentrated decontaminated sample using the reagents included in the kit according to the manufacturer’s instructions. Invalid assay results (absence of the human amplification control line) are usually obtained from specimens with a volume of < 200μl. Sample preparation of a set of 12 specimens takes approximately 45min. DNA amplification is performed with 10μl of disrupted sample supernatant and 15μl of the ready-to-use PCR mix. The amplification process is a multiplex PCR in which a triple target is amplified. DNA extraction is controlled by amplification of the human gene RNase P, which is also used to control for the presence of PCR inhibitors. The presence of Mycobacterium is assayed by amplifying a fragment of the 16S rRNA sequence and the presence of MTBC members by amplifying a fragment of the insertion sequence IS6110. DNA amplification was performed using a standard thermocycler and took 1h to complete. A dipstick with probes bound to colloidal gold and to the membrane is used for PCR product detection (Figure 1).
Figure 1:The PCR product is denatured for 2min and added to a preheated running solution in a thermoblock. After introduction of the dipstick and a 5min incubation period, the result is obtained from the presence of visible lines by using an automatic reading system. In our study, the results interpretation was performed automatically by the software according to the intensity of the line, but a visual interpretation is also possible based on the presence or absence of the red color on control and test lines.

Table 1:The positivity rate of the SO-DMT in smear positive and smear negative sputum samples when culture results were taken as gold standards.
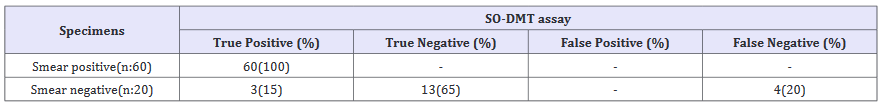
Table 2:The sensitivity, specifity, PPV and NPV of SO-DMT assay in smear positive and smear negative sputum samples culture results were taken as gold standard.

APositive predictive value, BNegative predictive value
The dipstick has four lines: a product control line, a PCR amplification control line, a test line for the whole Mycobacterium genus, whose presence in the absence of the M. tuberculosis complex line indicates the presence of NTM genetic material, and a test line for MTC members [14].
Results
All results were compared with Löwenstein Jensen culture. All smear positive sputum samples were positive with culture and Speed-oligo® Mycobacteria. Of 20 smear negative sputum samples, 7 were culture positive. Of 7 culture positive samples, 3 were positive SO-DMT (Table 1). SO-DMT assay showed no reactivity in any of the mycobacterium-free specimens or in those with 18 NTM reference strains. When LJ culture results were accepted as a reference method, the sensitivity rate of SO-DMT was 100 % for smear positive specimens and 42% for smear positive specimens (Table 2).
Discussion
TB is still one of the most contagious and deadly infections with approximately a quarter of cases not being identified and/or treated mainly due to a lack of resources. In the definitive diagnosis of tuberculosis, smear staining from clinical specimens and culture is gold standard. However, the sensitivity of the microscopy test is low. Culture requires an incubation period of 2-8 weeks for definitive diagnosis [15-19]. In general, patients with negative sputum microscopy at the first examination are being treated until the end of culture. Due to the contamination of the tuberculosis bacillus by droplets, patients with negative sputum microscopy may become infected during this period. These uncertainties in the diagnosis and treatment of tuberculosis play an important role in spreading the disease and worsening the disease in the patient [1-8]. The incidence of tuberculosis is increasing all over the world. Due to the increasing prevalence of multiple germline resistant strains and the emergence of a variety of immunosuppressive diseases such as AIDS, the use of faster and more sensitive diagnostic methods in the early diagnosis of this disease has become compulsory [20-22].
For this purpose, PCR - based automated and semi-automated kit based PCR systems developed specifically for MTBC diagnosis are widely used in routine diagnostic laboratories. The sensitivity and specificity of these methods vary according to the system used [6]. In a various studies, lung-derived specimens were analyzed by FDA approved Cobas Amplicor MTB (CAMPMTB; Roche Diagnostic Systems, Inc., Branchburg, NJ) and Amplified Mycobacterium tuberculosis Direct Test (AMTDII; Gen-Probe, Inc., San Diego, Calif. The sensitivity of these tests were between 55% and 97% and the specificity between 92% and 100% [6,8,10]. In our study, 60 sputum samples (40 smear positive, 20 smear negative) in smear positive and negative sputum samples collected from patient with suspected TB, 20 type reference strains of mycobacteria (18 non-tuberculosis mycobacteria, M. tuberculosis H37Rv and M. tuberculosis H37Ra) were evaluated with SO-DMT to determine sensitivitiy and specifitiy. When LJ culture results were accepted as a reference method, the sensitivity rate of SO-DMT was 100 % for smear positive specimens and 42% for smear positive specimens. The sensitivity of the assay was very satisfactory in AFB smear-positive specimens and correctly classified samples as MTBC or NTM. The sensitivity was lower (42%) in AFB smearnegative samples within the range previously reported for smearnegative samples [6,22]. The overall specificity was very high (100%). Several commercially available nucleic acid amplification tests have been extensively evaluated for the detection of MTBC species in respiratory samples, showing high specificity and good positive predictive values for smear-positive samples and variable sensitivity for smear-negative and extrapulmonary TB samples [6]. However, most of these tests require the use of real-time PCR or precision instruments that are not affordable in many countries with a high TB burden, or they involve technologies that require extensive hands-on time. In contrast, the SO-DMT assay combines a PCR amplification method with a simple detection procedure that does not require long post amplification preparation or exclusive equipment, reducing manipulation to only two pipetting steps. As limitations, assay automation is not possible and separate laboratory areas should be used for specimen preparation, target amplification, and amplicon detection.
In conclusion, the SO-DMT assay requires only 30min of handson time and offers a total turnaround time of 2h from sample reception. This method directly detects Mycobacterium species in a clinical specimen and simultaneously differentiates them from MTBC species. Results obtained in a convenience sample of AFB smear-positive specimens suggest that utilization of this assay for MTBC/NTM detection and differentiation can avoid the need to wait for culture results from smear-positive samples. This is an important benefit for case management and contact investigation, making the assay especially valuable for clinicians in countries with a high incidence of NTM [23,24]. SO-DMT is effective to identify for Mycobacterium tuberculosis complex and differentiation from genus mycobacterium in smear positive sputum samples. In this study, SO-DMT is not effective for diagnosis in smear negative sputum samples of MTBC. Wide range prospective study must be designed in smear negative and smear positive sputum samples collobration with clinical data.
References
- (1990) Diagnostic standards and classification of tuberculosis. American Thoracic Society Am Rev Respir Dis 142: 725-735.
- Ryu YJ, Koh WJ, Daley CL (2016) Diagnosis and treatment of nontuberculous mycobacterial lung disease: Clinicians perspectives. Tuberc Respir Dis 79(2): 74-84.
- Kendall BA, Winthrop KL (2013) Update on the epidemiology of pulmonary nontuberculous mycobacterial infections. Semin Respir Crit Care Med 34(1): 87-94.
- Prevots DR, Marras TK (2015) Epidemiology of human pulmonary infection with nontuberculous mycobacteria: A review. Clin Chest Med 36(1): 13-34.
- (2013) International Standards for Tuberculosis Care.
- Tarhan G, Cesur S, Şimşek H, Ceyhan İ, Özay Y, et al. (2015) Diagnostic value of nine nucleic acid amplification test systems for Mycobacterium tuberculosis complex. Journal of Microbiology and Infectious Diseases 5(3): 103-109.
- Waard JH, Robledo J (2007) Conventional diagnostic methods. In: Palomino JC, Leão SC, Ritacco V (Eds.), Tuberculosis 12: 401-424.
- Centers for Disease Control and Prevention (CDC) (2000) Update: nucleic acid amplification tests for tuberculosis. MMWR Morb Mortal Wkly Rep 49(26): 593-594.
- Hofmann Thiel S, Turaev L, Alnour T, Drath L, Müllerova M, et al. ( 2011 ) Multi-centre evaluation of the speed-oligo Mycobacteria assay for differentiation of Mycobacterium spp. in clinical isolates. BMC Infect Dis 11: 353.
- Lara OA, Mendoza LP, Rodriguez GJ, Fernández SAM, Bermúdez RMP, et al. ( 2013 ) Evaluation of the Speed-oligo Direct Mycobacterium tuberculosis assay for molecular detection of mycobacteria in clinical respiratory specimens. J Clin Microbiol 51(1): 77-82.
- Kubica GP, Dye WE, Cohn ML, Middlebrook G (1963) Sputum digestion and decontamination with N-acetyl-Lcysteine- sodium hydroxide for culture of mycobacteria. Am Rev Respir Dis 87: 775-779.
- Tuberculosis Division, International Union Against Tuberculosis and Lung Disease (2005) Tuberculosis bacteriology- priorities and indications in high prevalence countries: position of the technical staff of the Tuberculosis Division of the International Union Against. Int J Tuberc Lung Dis 9(4): 355-361.
- Tan WY, Stratton CW (2006) Diagnosis of Mycobacterium tuberculosis. Advanced Techniques in Diagnostic Microbiology, (2nd edn), Springer Newyork Heidelberg Dordrecht, London, UK, p. 567.
- http://www.vircell.com
- World Health Organization (2008) Global tuberculosis control: surveillance,planning, financing, Genava, Switzerland, 1-2.
- Gebre N, Karlsson U, Jönsson G, Macaden R, Wolde A, et al. (1995) Improved microscopical diagnosis of PTB in developing countries. Trans Royal Soc Trop Med Hyg 89(2): 191-193.
- Metchock BG, Nolte FS, Wallace Jr RJ (1997) Mycobacterium. In: Murray PR, Baron EJ, Pfaller MA, et al., (Eds.), Manual of clinical microbiology (7th edn), American Society for Microbiology, Washington DC, USA, 1999; 399-437. World Health Organization.Tuberculosis Diagnostic Workshop: Product Development guidelines, p. 21-24
- World Health Organization (1998) Laboratory services in tuberculosis control part III culture. Global Tuberculosis Programme, Geneva, Switzerland, pp. 47-52.
- Lambi EA (1993) Medium selection and incubation for the isolation of Mycobacteria. In: Isenberg HD (Ed.), Clinical microbiology procedures handbook. Volume 1, American Society for Microbiology, Washington DC, USA, pp. 3.6.1-3.6.8.
- Global Tuberculosis Report (2015) WHO/HTM/TB, p. 22.
- T.C.Sağlık Bakanlığı Verem Savaş Daire Başkanlığı, Türkiye’de Verem Savaşı 2017 Raporu 5-9.
- World Health Organization (2008) Global tuberculosis control: surveillance,planning, financing, Geneva, Switzerland, p. 1-2.
- Quezel Guerraz NM, Arriaza MM, Avila JA, Sanchez WE, Martinez MJ (2010) Evaluation of the Speed-oligo mycobacteria assay for identification of Mycobacterium spp. from fresh liquid and solid cultures of human clinical samples. Diagn Microbiol Infect Dis 68(2): 123-131.
- Wu X, Zhang J, Liang J, Lu Y, Li H, et al. (2007) Comparison of three methods for rapid identification of mycobacterial clinical isolates to the species level. J Clin Microbiol 45(6): 1898-1903.
© 2018 Gülnur Tarhan. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






