- Submissions

Full Text
Cohesive Journal of Microbiology & Infectious Disease
Molecular Detection of White Spot Syndrome Virus from Farmed Shrimps in Saudi Arabia
Alaudeen Hakami*
King Abdulaziz University, Saudi Arabia
*Corresponding author:Alaudeen Hakami, King Abdulaziz University, Jeddah, Saudi Arabia
Submission: April 02, 2018; Published: May 01, 2018

ISSN 2578-0190 Volume1 Issue4
Abstract
Shrimp industry is one of the important sectors in the kingdom of Saudi Arabia and it has shown steep production and economic values. WSSV is a major pathogen which causing 100% mortality and economic losses in world wide. In KSA two most important species such as Litopenaeus vannamei and Fenneropenaeus indicus were cultured widely. Due to the sheer expansion of shrimp culture industry, WSSV occurrences was found in F. indicus and followed by WSSV outbreak, SPF L. vannamei was imported and cultured in KSA with stringent biosecurity measures implied by ministry of water, environment and agriculture. The present study was aims to identify the presence of WSSV in L. vannamei in selected area of KSA applying molecular method. Samples were collected from Jazan region it was fixed in 95% ethanol and stored in -80°C for the analysis. The collected samples were subjected to nucleic acid (DNA) extraction, followed by conventional and real time PCR. The obtained positive PCR products were purified, and it was sequenced. The sequences analyzed to see the similarities in NCBI blast. The present study found that the collected samples were shown a clear positive in F. indicus and L. vannamei in conventional and real time PCR analysis. The obtained sequences were shown 97% similarities with other WSSV isolates in Gen Bank. The obtained data from this study was found to be the presence of WSSV in KSA. PCR followed by sequencing was useful to identify the presence of WSSV in a shrimp project area. Furthermore, we suggest more molecular studies for prevalence of WSSV.
Introduction
The aquaculture in Saudi Arabia was feasible due to weather condition that allow culturing different subtropical and tropical species. In addition, it has unique location surrounding by red sea to the west with wide tracts of land and the persian gulf coasts to the east and comprise 80% of the Arabian Peninsula that provide an excellent locality for the sea food harvest. So, lack of fresh water makes mariculture is the best option to supply increasing demand for the sea food in Saudi Arabia [1]. Since, 1970 when the Ministry of Agriculture in cooperation with Food and Agriculture Organization (FAO) have established Jeddah Fisheries Research Centre were identify the potential species suitable in fresh and marine water in Saudi Arabia and the aquaculture industry was encouraged in the 1980s by providing governmental subsidies to the farmers [2]. Nile tilapia and sigan have been the first produced in aquaculture until 2000 when the shrimp produced in large quantities and planned to produce up to 100, 000 tons/year [3]. The Indian white shrimp Fenneropenus indicus was found to survive and grow well in the high saline water around the country. Saudi Arabia have an ambitious strategy (Vision 2030) by investing more in aquaculture as part of an economic plan aim to reduce the dependence on oil. The aquaculture in Saudi Arabia is an important industry where recently report from FAO stated the Ministry of Environment, Water and Agriculture in Saudi Arabia has invested 10.6 billion into aquaculture projects with the purpose to produce one million tonnes of fish in the next years (Chalil, 2012). In 2017 an updated report for FAO account the capture of fishery production of the Saudi has been average of 68 000 tonnes during the last two years. Shrimps, mackerels, emperor fish and swimming crab are the main species caught [4]. In Saudi Arabia, there are 125 aquaculture farms and 56 of which are fully operational [5]. Worldwide, the major viral disease infecting shrimp are White Spot Syndrome Virus (WSSV). However, WSSV in Indian white shrimp (F. Indicus) were reported in Saudi Arabia [6]
and their origins, its virulent genotypes were determined [7,8]. Due to this WSSV outbreak the aquaculture production was decreased from 26 000 tonnes in 2009 and 2010 to 1000 tonnes in 2013 (Luis et al., 2016). As a result, all infected shrimp farming projects were closed [1]. In 2012, the ministry of environment, water and agriculture has perceived an idea to replace the currently cultured Indian white prawn F. indicus to the culture of Pacific white shrimp L. vannamei (specific pathogen free) they decided to import the L. vannamei (SPF) to Saudi Arabia [9]. The extensive studies were performed for environmental conditions, cohabitation, and feeding trials and samples were sent to the reference lab at Arizona University, and the results were negative for all the OIE/ USMSFP listed pathogens of shrimp and in 2014 all the shrimp farms changed culture from F. indicus to L. vannamei (Luis et al., 2016). The main aims of the present study were to screening for the presence of WSSV in cultured shrimp species L. vannamei (SPF) by molecular based methods (conventional PCR and confirmed by RT-PCR). The objective of this research was to identify the presence of WSSV in L. vannamei in selected area of Saudi Arabia applying molecular method
Materials and Methods
Samples were collected from Jazan region, fixed in 95% ethanol and stored in -80 °C for the analysis. The collected samples were subjected to nucleic acid (DNA) extraction, followed by conventional and real time PCR. The obtained positive PCR products were purified and it was sequenced. The sequences analysed to see the similarities in NCBI blast.
Sample collection
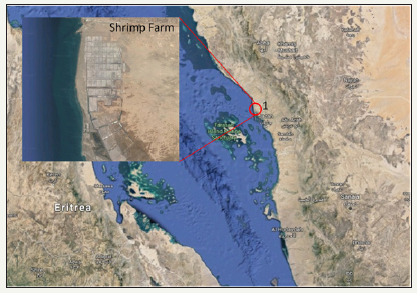
The samples used in the present study were obtained from moribund shrimps (Fenneropenaeus indicus and Litopenaeus vannamei) cultured in Jazan region, Saudi Arabia during epizootics in October, 2014 and August 2015 (Figure 1). The moribund shrimps were collected and dissected the pleopods at the farm site and fixed samples were transported to Fisheries Research Center, Jeddah, and it was stored in -80 °C for the downstrem apllication [10,11].
Figure 1: Location of the sample collection site Jazan, Saudi Arabia. 1. 17°15’11.2”N 42°20’52.5”E.
DNA extraction
The DNA extracted from fixed shrimp’s tissue samples using DNeasy Blood & Tissue Kit (Qiagen) following the manufacturer instrucuion. Briefly, the ethanol fixed pleopod samples were dried and incised in to small pieces. Aproximately 25mg of tissues was placed in 1.5ml microcentrifuge tube with 180μl Buffer ATL and 20μl proteinase K was added and it was mixed thoroughly. Smaples were mixed till the complete mixing and 20ul of AL buufer was added, followed by 2nd round of mixing. Next, 200μl of 100% ethanol was added in the reaction and mixed again. Sample was loded on DNeasy Mini spin column which was kept in 2ml collection tube. The tubes were centrifuged in 8000rpm for 1 minute and flow through was discarded and placed the DNeasy Mini spin column in a new collection tube. In the column 500μl Buffer AW2 was added, and centrifuged for 3min at 14,000rpm subsequently discarded flow-through and collection tube. Finally placed the DNeasy Mini spin column in a clean 1.5ml microcentrifuge tube, and 200μl Buffer AE was added directly onto the DNeasy membrane and kept in a room temperature for 1 minute. The elution was carried out with centrifuging the DNeasy Mini spin column at 8000rpm in 1 minute. The eluted DNA was assessed by spectrophotometer (Nanodrop 2000, Thermo Scientific) and its quality and quantity were determined.
Nested PCR assay for the Confirmation of WSSV
Table 1: Primers used for PCR analysis of WSSV confirmation
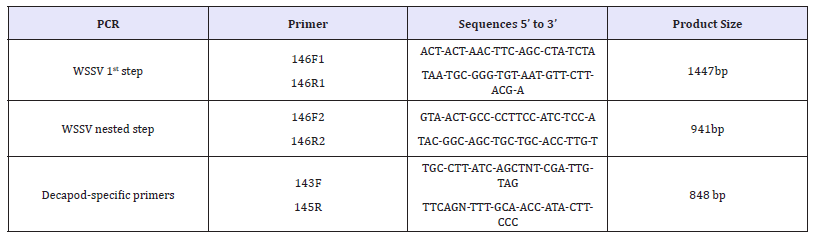
A nested PCR assay was developed for highly sensitive detection of WSSV. The extracted DNA was analyzed by nested PCR to confirmed the WSSV infection with the modified PCR program based on OIE recommendation [12,13]. The 25μL of PCR reaction mixture was prepared according to the manufacturer instructions (ThermoFisher Scientific, USA). Each PCR reaction contains 1μl (100ng) of extracted DNA, 2.5μl of 10X dream taq green buffer (ThermoFisher Scientific, USA ), 2.5μl of dNTPs mix, 1μl of each primer (5 micro moles), 0.4 μl of dream taq DNA polymerase enzyme and 16.6μl of nuclease free water. Thermal cycler conditions for the PCR reaction were as follows, initial denaturation at 94 °C for 3min, followed by 39 cycles of denaturation at 94 °C for 20 sec, annealing at 62 °C for 1 minute, extension at 72 °C for 1 minute and a final extension step at 72 °C for 3min. The product from the first step PCR was used as template for second step PCR. To verify the extracted DNA Decapod-specific primers used as control reactions [14]. PCR products were analyzed by 1.2% agarose gel electrophoresis and the gel was visualized under UV trans-illuminator. The primer sequence details are listed in the Table 1.
Results
Confirmation of DNA by internal control primer (Decapod-specific primers)
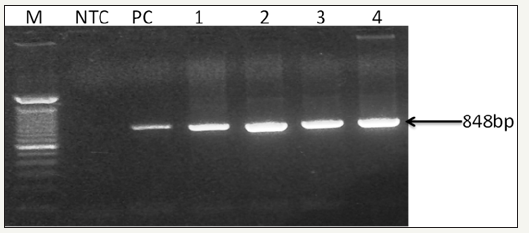
Internal Control Decapod-specific (18s RNA) primers (143F 5’-TGC-CTT-ATC-AGCTNT-CGA-TTG-TAG-3’ and 145R 5’-TTCAGNTTT- GCA-ACC-ATA-CTT-CCC-3’ yielding an 848 bp amplicon (Figure 2). In this study decaped specific primers were employed to determine the integrity and specificty of PCR. The 18 S RNA seuence was speficially ammplied in all PCR cases.
Figure 2: 0.8% Agarose gel photograph of PCR product, using Decapod-specific primers (OIE - Manual of Diagnostic Tests for Aquatic Animals, 2017) for the detection of virus in test and control Shrimp samples DNA. Captions: LaneM, stander size molecular weight (100bp DNA ladder, Cat No; 15628019, Thermo Scientific, CA), LNTC, Non- template Control, L PC, Positive Control, L1-4, F. indicus and F. indicus test samples..
Detection of WSSV by nested PCR (OIE)
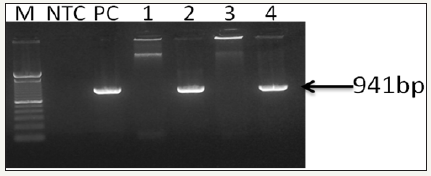
The extracted DNA was eluted and stored at –20 °C. The DNA purity and concentration was determined by agarose gel electrophoresis and spectrophotometer. The first-step and the second step (nested) PCR agarose gel electrophoresis showed different bands (Figure 3 & 4).
Figure 3: 0.8% Agarose gel photograph of first-step PCR for the confirmation of WSSV (OIE - Manual of Diagnostic Tests for Aquatic Animals, 2017). Captions: Lane M, stander size molecular weight (100bp DNA ladder, Cat No; 15628019, Thermo Scientific, CA), LNTC, Non template Control, LPC, DNA Positive Control, L1, F. indicus negative control, L2, F. indicus positive, L3, L. vannamei SPF, L4, L. vannamei positive..

Figure 4: 0.8% Agarose gel photograph of second step (nested) PCR agarose gel electrophoresis for the confirmation of WSSV (OIE - Manual of Diagnostic Tests for Aquatic Animals, 2017). Captions: Lane M, stander size molecular weight (100bp DNA ladder, Cat No; 15628019, Thermo Scientific, CA), LNTC, Non- template Control, LPC, DNA Positive Control, L1, F. indicus negative control, L2, F. indicus positive, L3, L. vannamei SPF, L4, L. vannamei positive.
Detection of WSSV by RT-PCR (OIE)
The presence of WSSV in F. indicus and L. vannamei samples from naturally infected shrimps in Jazan area, KSA, is depcited in the standard curve (Figure 5). A ten-fold serially diluted bp WSSV DNA fragment was amplified and analyzed in real-time. The standard curves and quantification cycle (Cq) values for quantitative realtime PCR are plotted against copy number as shown (Figure 6) Table 2.
Figure 5: Real time graph showing the amplification of WSSV from tested samples (F.indicus and L.vannamei
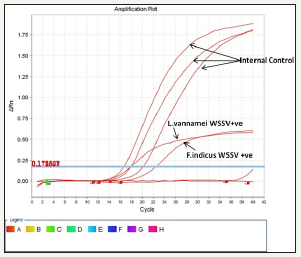
Figure 6: Nucleotide sequence of the WSSV genes amplified in this study.

Table 2: Conidtions (Target, Reporter and Quencher) used for the amplification and confirmation of WSSV from F.indicus and L.vannamei by IQ 2000 real time kit method.
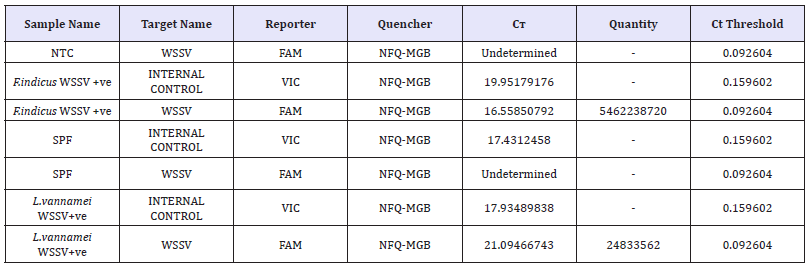
BLAST analysis (Table 3 & 4) (Figure 7 & 8)
Table 3:The Nucleotide similarity and identity of WSSV isolate from F.indicus showed in BLAST analysis.
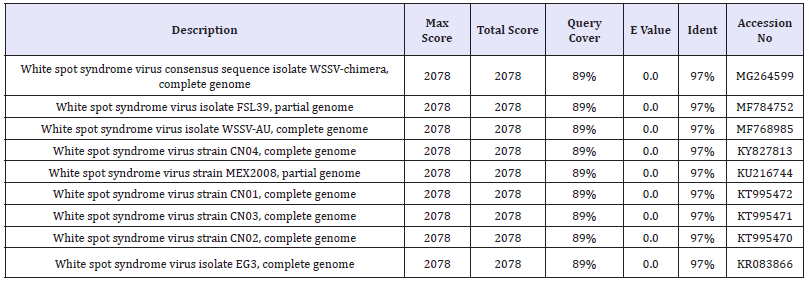
Table 4: The Nucleotide similarity and identity of WSSV isolate from L.vennamei showed in BLAST analysis.
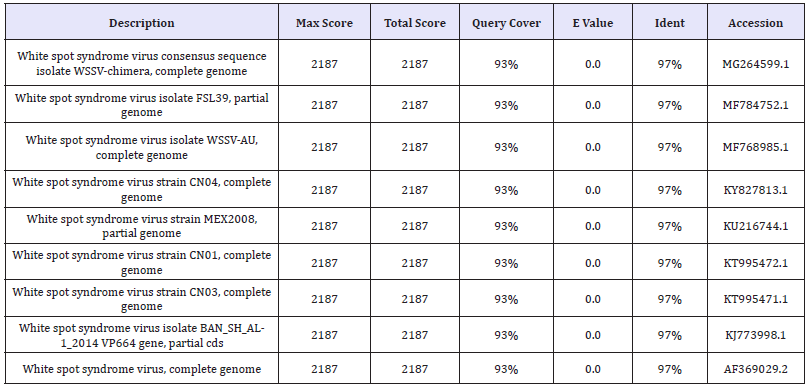
Figure 7: Translated AA sequences from amplified WSSV gene from F.indicus.

Figure 8: Translated AA sequences from amplified WSSV gene from L.vennamei.

Discussion
Worldwide, white spot syndrome virus (WSSV) is considered one of the most pathogenic and devastating threat to the commercial shrimp farming. In addition, the virus affect other fresh and marine crustaceans such as Crayfish, Crabs and lobsters. In fact, WSSV is highly virulent in shrimp farms, particularly in shrimp culture type within few days it can spread leading to 100% mortality. In Saudi Arabia, the WSSV was first reported in 2010 affected cultured F. indicus species and resulting in massive economic losses (Kathy et al., 2012). To date, several reports have been published describing the economic losses of WSSV in Saudi Arabia.
Ministry of Environmental, Water and Aquaculture as longterm solution instead F. indicus infected species to SPF L. vannamei resistant to WSSV with offers many benefits to shrimp industry. Epidemiological studies and precise information on the economic impact of WSSV throughout the country is not available and is required to overcome the expected disaster and disease outbreaks. The results of the present study showed a clear WSSV DNA positive band from F. indicus and L. vannamei samples by conventional PCR. The positive samples were further detected and confirmed by RTPCR analysis. The detected positive WSSV in the samples were sequenced and were showed 97% similarities with other WSSV sequences published in GenBank. Contrary to the previous believes, the presence of WWSV was confirmed in KSA. However, the result could postulate that the WSSV is more common in KSA than previously thought and could have economic impacts on the local investment. The virus could introduce to the country through the imported shrimps (Kathy et al., 2012). The result of the examined samples by RT-PCR, could indicate that the current technique is robust, sensitive and highly specific molecular technique for the detection of WSSV therefore, it is recommended to detect the virus in infected shrimp at field level.
Conclusion and Recommendations
The analysed data from the current study indicate the infectivity of WSSV to the shrimps in KSA. Conventional, real time PCR followed by sequencing was potent to clarify the presence of WSSV in shrimp cultivated area given an alarming for industrial disaster. Based on our findings of this study, we conclude that PCR based technique is a good measure for accurate measurement of DNA molecule in sequence specific manner. Furthermore, we suggest more molecular-based studies for prevalence WSSV among shrimp farms in KSA to identify the risk factors of the coming disease outbreaks.
Acknowledgment
We thank KACST, Riyadh and DSR, KAU for the financials support. We are very grateful to the Director Jeddah Fisheries Department for allowing the use of their facilities in perusal of these goals. Special thanks to Miss Mira for her kind help.
References
- Nord Oest (2016) Aquaculture in Saudi Arabia. Innovasjon Norge, Norway, pp. 1-53.
- FAO Fisheries statistics of Saudi Arabia (2006) Marine Fisheries Department, Ministry of Agriculture, Kingdom of Saudi Arabia.
- http://www.fao.org/in-action/globefish/fisheryinformation/resourcedetail/ en/c/338614/
- FAO Fisheries statistics of Saudi Arabia (2003) Marine Fisheries Department, Ministry of Agriculture, Kingdom of Saudi Arabia.
- Ming-hsien Lin (2001) ICDF- Aquaculture development in the kingdom of Saudi Arabia. International Cooperation & Development Middle East, pp. 20-24.
- OIE (Office International des Epizooties/World Animal Health Organization) (2011a) Immediate notification, submitted date: 04/10/2011. OIE, Paris.
- Tang KF, Navarro SA, Pantoja CR, Aranguren FL, Lightner DV (2012) New genotype of white spot syndrome virus (WSSV) and Taura syndrome virus (TSV) from the Kingdom of Saudi Arabia. Dis Aquat Organ 99(3): 179-185.
- Tang KF, Pantoja CR, Redman RM, Lightner DV (2012) A histological variant of white spot syndrome virus (WSSV) from the Kingdom of Saudi Arabia. Journal of Invertebrate Pathology 113(1): 82-85.
- Aranguren FL (2016) Saudi Arabia developing effective farmed shrimp biosecurity strategy. Global Aquaculture Alliance.
- Nord Oest (2016) Aquaculture in Saudi Arabia. Innovasjon Norge.
- Arthur JR, Alday-Sanz V, Doyle RW, Mather PM, Hurwood D, et al. (2012) Proposal to introduce whiteleg shrimp (Litopenaeus vannamei) to the kingdom of Saudi Arabia for aquaculture development. Prepared for Saudi Aquaculture Society.
- Chu-Fang Lo, Jiann-Horng Leu, Ching-Hui Ho, Chau-Huei Chen, Shao-En Peng, et al. (1996) Detection of baculovirus associated with white spot syndrome (WSBV) in penaeid shrimps using polymerase chain reaction. Dis Aquat Org 25: 133-141.
- Nunan LM, Lightner DV (2011) Optimized PCR assay for detection of white spot syndrome virus (WSSV). J Virol Methods 171: 318–321.
- OIE - Manual of Diagnostic Tests for Aquatic Animals, CHAPTER 2.2. 8. White Spot Disease-2017.
© 2018 Alaudeen Hakami. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






