- Submissions

Full Text
Biodiversity Online J
Comprehensive Evaluation of Quercus floribunda (Oak): Insight from Molecular Markers and Phylogeny
Ayesha A1, Abdul M1*, Khushi M2, Sajjad2, Daniel P3, Hamid A2 and Irfan U2
1Department of Botany, Hazara University Mansehra, Pakistan
2Department of Biotechnology and Genetic Engineering Hazara University Mansehra, Pakistan
3Department of plant sciences, University of California UC Davis, USA
*Corresponding author:Sajjad, Department of Biotechnology and Genetic Engineering Hazara University Mansehra, Mansehra 21300, Pakistan
Submission: April 16, 2025; Published: July 08, 2025

ISSN 2637-7082Volume5 Issue 3
Abstract
The Quercus L. (Fagaceae) represents a complex taxonomic genus with approximately 600 extant species of significant ecological and economic importance. Accurate species identification remains challenging due to frequent hybridization and morphological similarities. This study influences DNA barcoding as a powerful molecular approach to distinguish Q. floribunda L., a species native to the Himalayan regions of Pakistan. In the current study, three chloroplast DNA barcoding loci, the rpoB, rbcL and tRNA, were investigated to assess their effectiveness in Q. floribunda L. identification and Phylogenetics. The research employed comprehensive molecular techniques to evaluate genetic diversity and evolutionary relationships of Q. floribunda L. The rpoB gene revealed the highest discriminatory power, exhibiting superior haplotype diversity, and provided insights into relationships between Q. floribunda L. and other Quercus species, with rpoB showing 14 haplotypes, 5 rbcL, and 8 tRNA. Phylogenetic analysis placed Q. floribunda L. in a shared ancestral node with Q. variables for rpoB, while rbcL showed high similarity with Q. aucheri. The tRNA spacer revealed evolutionary divergence across Quercus species, classifying sequences into four major clades. In conclusion, all the barcode regions demonstrated considerable diversity among Quercus species, supporting their utility as barcodes for identification. This barcoding study represents the first comprehensive analysis on Quercus from the Himalayan region of Pakistan. The findings contribute to accurate species identification, phylogenetic understanding, and conservation efforts for this ecologically important oak species. This research also highlights the potential of DNA barcoding in resolving taxonomic uncertainties and its application in biodiversity conservation and management of Quercus and other valuable plant resources.
Keywords:Genetic distance estimation; Quercus; Universal markers; rpoB; rbcL; tRNA; Evolutionary relationship
Introduction
Quercus L. (Fagaceae) is an evolutionarily diverse genus and comprises about 600 species worldwide [1]. This genus is considered the most widely circulated woody genus in the northern hemisphere [2,3]. Q. floribunda L. is commonly known as holly oak and locally known as bunj or barungi [4,5]. This tree grows up to 20-22 meters tall. The leaves are typically elliptic-ovate to broadly lanceolate and are leathery (coriaceous) with entire or spiny toothed edges [6]. Both sides of the leaves are smooth and green (glabrous), with an oblique base. The petiole is 0.3 to 1cm long [7]. This species is differentiated from other members of the genus by its bicolored green leaves [8]. April to May is noted for the flowering period of Q. floribunda L. Sometimes, Q. floribunda L. and Q. baloot are observed as hybrids resembling those featuring leaves [9]. These hybrids may result from cross-pollination between Q. floribunda L. and Q. baloot [10]. Quercus species are primarily distributed in the temperate and sub temperate areas of the western and northern parts of Pakistan [11]. These areas are the Himalayan foothills, Karakoram and Hindu Kush ranges, such as Azad Jammu and Kashmir, Khyber Pakhtunkhwa (particularly Abbottabad, Mansehra, Chitral, Dir and Swat), Gilgit-Baltistan, and the high- altitude zones of Baluchistan, including the Suleiman Range [12]. Q. floribunda L. contains a variety of phytochemical constituents such as flavonoids (kaempferol and quercetin), phenolic acids (ellagic and gallic acids), lignans, sterols (β-sitosterol), tannins (both condensed and hydrolysable), triterpenes (oleanolic and ursolic acids), and volatile oils [13]. These compounds play a crucial role by exhibiting the activities such as anti-inflammatory, antidiabetic, antioxidant, anticancer and antimicrobial effects [14,15].
Due to hybridization and molecular evolution, oak trees exhibit sympatric parallel diversification accompanied by leaf morphological variations and harbor a high potential for introgression and reticulate evolution [16]. Such evolutionary divergence and similar morphology also pose a challenge in identifying members of the Fagaceae [17]. Conversely, molecular and DNA diagnostic tools such as plastid DNA barcodes and multilocus DNA markers can identify [18]. DNA barcoding is an advanced technological approach used to identify unknown biological taxa at the genus, species, or even subspecies level with high accuracy [19,20]. DNA barcoding aims to accurately determine plant species by analyzing a specific DNA fragment within a short timeframe. This method is recognized for its efficiency, standardization, speed, and cost-effectiveness in identifying diverse plant species [21]. This technique permits the investigation of both intra-specific and interspecific variations, facilitating the identification of unknown species or those with complex morphometric traits [22]. DNA barcoding markers, comprising rpoB, rbcL, ITS, and tRNA, have been used to identify plant species [23]. These markers play a critical role in examining the similarities and differences between various species, as well as in addressing taxonomic and evolutionary challenges [24]. Numerous plastid genomic loci, comprising rpoB, rpoC1, rbcL, and tRNA, have been utilized as DNA barcoding for plant identification [25-27]. The present study aims to assess the Quercus floribunda population from Northern Pakistan using molecular markers to estimate genetic distances and establish phylogenetic relationships with reference sequences from databases.
Materials and Methods
Sample collection and identification
Q. floribunda L. samples were collected and studied from natural habitats of Northern, Pakistan. Detailed information about the species were presented in the (Table 1). The collected samples were identified by the expert taxonomists and preserved at the Herbarium of Hazara University Mansehra, Pakistan.
Table 1:Detailed description of selected species along with primary data in the form of GPS coordinates and collection site.

DNA extraction and PCR amplification
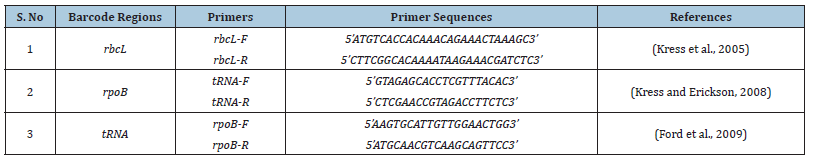
For DNA extraction and PCR amplification, whole genomic DNA was extracted using the CTAB method, described by [28], with some optimizations. The extracted DNA samples were quantified using 1% agarose gel electrophoresis. Three genetic markers were amplified: the rpoB gene, rbcL gene, and tRNA barcodes, which were recommended by the Consortium for the Barcode of Life (CBOL) [29]. The primer pairs used for PCR amplification are listed in Table 2. The extracted DNA was stored at -20 °C and utilized as a template for amplifying barcode genes and spacers. The PCR reaction mixture (25μL) comprised 2.5μL of 10×PCR buffer (without MgCl2), 2μL of each dNTP, 1.25μL of each primer, and 0.125μL of Taq DNA polymerase. PCR reactions were carried out at 95 °C for 5 minutes, followed by 35 cycles of 94 °C for 30 seconds, 52 °C for 30 seconds, and 72 °C for 45 seconds. The final extension step was at 72 °C for 10 minutes. Macrogn Inc. performed sequencing in Korea, and all resultant sequences were submitted to the DNA Data Bank of Japan (DDBJ) for accession numbers.
Table 2:Detailed description of selected DNA barcode regions it’s with forward and reverse primers along with recommended authorities.
Nucleotide sequences alignments and analysis
After successful sequencing of the rpoB, rbcL, and tRNA, the good-quality sequences were used to align in online NCBI GenBank tools to retrieve the most similar sequences clustered with Q. floribunda L. with its closest relative’s reference sequences and diverse species sequences. Generous software was used to check the quality of sequences, and missing sequences, whether at the start or the end, were carefully reviewed and deleted, ensuring our data’s highest quality and reliability. The query sequences along with reference sequences were used for multiple sequence alignment in Clustal W [30]. Phylogenetic analysis was performed to identify the relationship between individuals, species, and genus. Neighbour-joining trees were constructed through Cantor- Jukes model with 10,000 bootstraps. The other analysis, such as nucleotide diversity (π), segregation (S), haplotype diversity (Hd), haplotype diversity distribution, and haplotype diversity network, were conducted in Python version 3.10.10.
Results
Molecular analysis of Quercus floribunda
Table 3:A detailed description of DNA barcodes sequences submitted into NCBI (DDJB) database along with obtained Accession IDS.
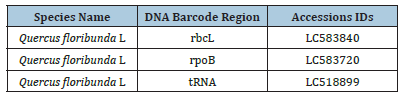
In this study, three barcode regions, such as rbcL, rpoB, and tRNA, were selected and successfully amplified using forward and reverse primers. The initial identification of Quercus spp. was performed through morphological techniques and tagged as Q. floribunda L. The sequencing results of these barcodes were submitted to the NCBI GenBank, and obtained the accessions LC58840, LC583720, and LC518899 for rbcL, rpoB, and tRNA (Table 3). In the current study nucleotide diversity, evolutionary relationship, phylogeny, and haplotype diversity analysis Q. floribunda L were performed.
Phylogeny and evolutionary analysis of Quercus floribunda through rpoB marker
Current study assessed the genomic similarity, alignment, and phylogenetic trees based on rpoB, rbcL, and tRNA. Besides the reference rpoB (LC583720), 24 sequences were detected with a high similarity threshold via Mega Blast. Apart from L. craibianus and L. litseifolius, the dataset contains 22 Quercus spp sequences. The genetic similarity level of Q. floribunda L. was 1-2, which was higher as compared to other Quercus spp (Figure 1a). It has been mentioned that oak species are prone to molecular evolution despite having higher morphological similarities. Based on the phylogenetic tree, Q. floribunda shared an ancestral node with Q. variables. However, Q. chenii, Q. acutissima, Q. serrata, and Q. baronii shared the same clade, and presented in (Figure 1b).
Figure 1:Analysis of the rpoB gene-aligned sequences.
a) Indicates a 2D matrix of the pairwise-sequence distances. Pairwise distances are represented with color schemes, from dark
blue (lower) to yellow (higher).
b) Phylogenetic tree classification of rpoB genes generated with 1000 bootstraps using Neighbor-Joining (NJ). Light blue circles
represent bootstrap confidence; the larger circle size corresponds to higher bootstrap confidence.

Phylogeny and evolutionary analysis of Quercus floribunda through rbcL
Based on rbcL sequence analysis, the query sequence LC58840 along with similar sequences were used to align. The pairwise sequence similarity indicated that the higher similarity of Q. baloot and Q. aucheri and divergence with Q. acutissima and Q. chenii, respectively. Phylogenetic analysis classified Q. floribunda closest to Q. baloot, Q. aucheri, and Q. coccifera among the Quercus spp presented in (Figure 2a & 2b). Phylogeny and evolutionary Analysis of Quercus floribunda through tRNA: Based on query tRNA sequence along with total of 14 highly similar Quercus spp sequences retrieved from GenBank were used for analysis. In the phylogenetic tree Q. floribundaQ. senescens, Q. semecarpifolia, tungmaiensis, Q. aquifolioides, Q. rehderiana, and Q. brandisiana, presented in (Figure 3a & 3b).
Figure 2:

Figure 3:Analysis of the tRNA spacer-aligned sequences.
a) indicates a 2D matrix of the pairwise-sequence distances. Pairwise distances are represented with color
schemes, from dark blue (lower) to yellow (higher)
b) Phylogenetic tree classification of tRNA genes generated with 10000 bootstraps using Neighbor Joining (NJ).
Light blue circles represent bootstrap confidence; the larger circle size corresponds to higher bootstrap confidence.

Assessment of diversity analysis
The assessment of nucleotide diversity (π), segregation sites (S), haplotype identification (Hd), distribution, and networking for the genomic aligned sequences of rpoB, rbcL, and tRNA spacer were performed. Guo and co-workers have suggested similar analyses for Theaceae plants. A designated Python script was used for calculating π using the sequence pairwise distances. Current study results indicated 0.092, 0.025, and 0.011 for rpoB, rbcL, and tRNA, respectively. Furthermore, to access the segregation site (S) in each dataset 338, 5, and 21 each for rpoB, rbcL, and tRNA were detected presented in (Figure 4). The segregation data S, and haplotype diversity (Hd), indicating 0.913 for (rpoB), 0.722 (rbcL), and 0.836 (tRNA) were recorded presented in (Figure 5). All these barcode regions exhibited higher haplotype diversity, and recommended as good barcode for species identification.
Figure 4:Calculation of the Segregation Sites: Segregating sites are calculated in Python version 3.10.10. Panel A, B, and C represent rpoB, rbcL, and tRNA, respectively.

Figure 5:Haplotype Distribution Analysis: The calculated haplotypes are displayed in a pie chart. Below, the memberships of each dataset are mentioned, with their respective representations in the analyses.

Analysis of haplotype diversity
Based on these barcodes sequences haplotypes and distribute their diversity (Hd) were calculated. The results indicated that 14, 5, and 8 haplotypes in the rpoB, rbcL, and tRNA, respectively. As expected, the genes retaining higher (Hd) scores generated more haplotypes and showed higher diversity. The results found that all the barcodes’ regions achieved considerable diversity among the Quercus spp. Furthermore, the relationship was calculated between these haplotypes. Thus, the networks were employed in Python and used hamming distances between the nodes (haplotypes) and applied them to edge weights, i.e., higher weight corresponds to more thickness to describe the distance between the haplotypes. Based on rpoB, the reference node (node 12) exhibited a connection with other haplotypes retaining different distances. Among them, haplotype 8 had the close relationship with Q. brandisiana. Similarly, the reference rbcL shares the same clade with Q. baloot and Q. aucheri. Based on tRNA haplotype network, the reference node shares a similar distance with other haplotypes 7 (Q. oxycodone and Q. kiukiangensis), and lowest distance of 13 haplotypes presented in (Figure 6).
Figure 6:Haplotype Distribution Analysis: The calculated haplotypes are displayed in a pie chart. Below, the memberships of each dataset are mentioned, with their respective representations in the analyses.

Discussion
The emergence of advanced sequencing technologies has significantly enhanced the utility of DNA barcoding in taxonomy, particularly for species identification and assessing biological diversity across populations and communities [20]. The current study highlighted the effectiveness of chloroplast regions such as rbcL, rpoB and tRNA recommended the Consortium of Life (CBOL) [29]. These markers have shown a reliable species identification and genetic diversity within various plant taxa, including oaks (Quercus spp.) [31,32]. The current study result is consistent with the previous research on Quercus species, which exhibit the high level of genetic variation due to their extensive distribution and ecological adaptability [33]. This genetic variability is essential for the Quercus species adaptation in diverse environmental conditions [34]. The high nucleotide diversity observed in the rpoB gene suggests significant evolutionary potential, which aligns with the previous studies that highlight the role of genetic diversity in facilitating the adaptive responses to environmental changes [35]. In the current study, phylogenetic analysis placed the Quercus floribunda L. in close relation to other members of the genus, such as Q. variabilis and Q. acutissima, which reflect the complex evolutionary history of oaks characterized by hybridization and gene flow [36,37]. These findings are strongly supported by interspecific hybridization events within the Quercus genus and contributes to their phylogenetic complexity [38]. Similarly, a previous study demonstrated that hybridization and introgression are common in the genus Quercus, blurring the species boundaries and complicating phylogenetic reconstructions [39]. On the other hand, similar findings were reported on Q. variabilis and Q. acutissima in East Asia, where overlapping habitats facilitated genetic exchange [40]. These patterns resonate with the placement of Q. floribunda alongside these species, suggesting that shared gene flow may strengthen their close genetic relationship. Similar results were found that Asian oaks often exhibit higher genetic diversity and reticulate evolutionary patterns [41]. Wang [11] also used ITS and chloroplast markers to resolve phylogenetic relationships among Asian oaks and found that Q. variabilis and Q. acutissima form a closely knit cluster. Studies on Quercus robur and Quercus petraea in Europe also highlight similar patterns gene flow and hybridization, showing that these processes are not confined to Asian species but represent a global phenomenon in the Quercus genus [42]. Understanding the genetic diversity and population structure of Q. floribunda L. is essential for conservation strategies because conservation efforts concentrate on preserving the genetic connectivity across landscapes [43]. This is a major step in ensuring the oak species long-term variability and adoptability [44]. Molecular markers such as rbcL, rpoB and tRNA genes provide a robust framework for assessing the genetic diversity and phylogenetic relationship [45]. Plant systematics have widely used these markers to resolve the taxonomic uncertainties and understand the evolutionary processes [46]. Integrating the chloroplast and nuclear DNA data enhances the resolution of phylogenetic analysis, offering a comprehensive view of species relationships within complex genera like Quercus [47]. The applications of these DNA barcodes have been further validated through comprehensive analysis, such as pairwise-sequence similarity tests, phylogenetic establishment and haplotype networking, which collectively establish their potential to resolve the taxonomic uncertainties and also understand the evolutionary relationship among species [48]. The current study focusing on the Quercus floribunda from the northern region of Pakistan and the finding of the results rpoB gene exhibited the highest haplotype diversity among the three tested barcode regions, indicating the superior capacity of the rpoB gene in distinguishing closely related species within the Quercus genus. Markers for terrestrial plants and underscored the importance of selecting appropriate markers for effective specieslevel identification [49]. The rapid evolution rate in the rpoB gene may contribute to its effectiveness in capturing recent speciation events, which is particularly relevant given the challenges posed by hybridization and introgression in oak taxonomy [50]. However, it is essential to recognize that relying only on a single barcode region may only sometimes yield sufficient resolution for all species within a diverse genus like Quercus [51]. Current study finding supports multi-barcode approaches to enhance species identification and their phylogenetic analysis, as supported by the literature and study of Heise [52] and Gonzalez [43]. Furthermore, current study haplotype network analysis revealed significant insight into the relationship among different Quercus species, suggesting recent divergence or ongoing gene flow, particularly between Q. floribunda and closely related species such as Q. baloot and Q. aucheri. This information is crucial for understanding the evolutionary history and biogeography of oaks in the region [53,54]. The observed genetic variability within Q. floribunda and its relative is vital for the adaptive potential of these species, especially in the context of climate changes and environmental pressures [55]. Conservation strategies should, therefore, consider this genetic diversity to preserve the evolutionary potential of these important forest trees [56-61].
Conclusion
In the current study, three barcode regions were successfully amplified, sequenced, and analyzed through various computational online and offline tools, both. The obtained sequences evaluation and identification were carried out using reference sequences of the Quercus genus retrieved from GenBank. The primers used in the current study showed over 90% effectiveness. The chloroplast markers, such rpoB, rbcL, and tRNA, performed exceptionally well in species identification and are recommended for use in evaluating other Quercus species across northern Pakistan and globally. Phylogenetic analysis showed that Quercus floribunda from the study area is most closely related to Q. baloot and Q. aucheri, and somehow with other species such as Q. variabilis, Q. tungmaiensis, Q. senescens, Q. semecarpifolia, Q. aquifolioides, Q. rehderiana, and Q. brandisiana. These findings lay the foundation for further research on the genetic diversity, population structure, and species identification within the Quercus genus.
Acknowledgement
All authors extend their sincere appreciation to the Researchers Supporting Project No: 5711 and the Department of Botany and Department of Biotechnology and Genetic Engineering, Hazara University Mansehra, KP, Pakistan, for assistance and technical support in the experiment.
Author Contribution
AA and AM conceived and designed the study. AA S and IU performed the experiments. AA, S, HA and IU contributed to data collection and analysis. AA, AM, S, DP and HA assisted in manuscript drafting and reviewing. AM KM and DP provided supervision and critical revisions. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Morales SS, Hipp AL, Valencia A, Hahn M, González MSE (2024) Divergence and reticulation in the Mexican white oaks: Ecological and phylogenomic evidence on species limits and phylogenetic networks in the Quercus laeta complex (Fagaceae). Annals of Botany 133(7): 1007-1024.
- Chen X, Li B, Zhang X (2023) Comparison of chloroplast genomes and phylogenetic analysis of four species in Quercus section Cyclobalanopsis. Scientific Reports 13(1): 18731.
- Shourmij M, Fard JK, Najafizadeh P, Mousavi Z (2022) Safety assessment of the Quercus brantii gall hydroalcoholic extract: Single and repeated oral dose toxicity studies. Iranian Journal of Basic Medical Sciences 25(11): 1389.
- Nisar MS, Wadood S, Iqbal A, Nausheen A, Ghafoor A (2016) Intra and inter specific profiling of Pakistani Quercus species growing in the hilly areas of district Dir Khyber Pakhtunkhwa. Pakistan Journal of Botany 48(1): 263-270.
- Akhtar MF (2020) HPLC analysis and pharmacological study of Quercus dilatata ex-Royle aqueous methanolic extract in Sprague Dawley rats. Pakistan Journal of Pharmaceutical Sciences 33(1): 1327-1332.
- Lemmens R (2023) Useful trees of East Africa: Uganda, Kenya, Tanzania.
- Muhammad N, Castillejo MA, Rey MD, Jorrín JVN (2023) An overview of oak species in Pakistan: Past, present, and future research perspectives. Forests 14(4): 777.
- Shah ST, Ahmad H, Zamir R (2005) Pollen morphology of three species of Quercus (Family Fagaceae). Journal of Agriculture, Forestry and the Social Sciences 1(4): 359-360.
- Denk T, Velitzelos D, Güner TH, Bouchal JM, Grímsson F, et al. (2017) Taxonomy and paleoecology of two widespread western Eurasian Neogene sclerophyllous oak species: Quercus drymeja Unger and Q. mediterranea Unger. Review of Palaeobotany and Palynology 241: 98-128.
- López DH, Vázquez FM, Soto A (2017) The role of hybridization on the adaptive potential of Mediterranean sclerophyllous oaks: the case of the Quercus ilex x suber complex. Oaks Physiological Ecology Exploring the Functional Diversity of Genus Quercus L pp: 239-260.
- Wang Y, Xu C, Qang W, Jiang Y, Qin L (2022) Germplasm resources of oaks (Quercus L.) in China: Utilization and prospects. Biology 12(1): 76.
- Ihtisham M (2013) HPLCDAD analysis and free radical scavenging potential of Quercus dilatata Pakistan Journal of Botany 45(S1): 577-581.
- Ahmad FM, Zafar AM, Ahmed NA, Hasan MMU (2023) Quercus floribunda ex a. camus: A tremendous remedy against inflammation and associated symptoms. Fitoterapia 170: 105-628.
- Taib M, Rezzak Y, Bouyazza L, Lyoussi B (2020) Medicinal uses, phytochemistry, and pharmacological activities of Quercus Evidence‐Based Complementary and Alternative Medicine 20(1): 1920683.
- Islam M, Ahmad I, Akhtar N, Alam J, Razzaq A (2021) Medicinal plants resources of Western Himalayan Palas Valley, Indus Kohistan, Pakistan: Their uses and degrees of risk of extinction. Saudi Journal of Biological Sciences 28(5): 3076-3093.
- Hipp AL, Manos PS, Gonzalez AR, Hahn M, Kaproth M (2018) Sympatric parallel diversification of major oak clades in the Americas and the origins of Mexican species diversity. New Phytol 217(1): 439-452.
- Fortini P, Marzio PD, Pietro RD (2015) Differentiation and hybridization of Quercus frainetto, petraea, and Q. pubescens (Fagaceae): Insights from macro-morphological leaf traits and molecular data. Plant Systematics and Evolution 301: 375-385.
- Showalter M, Sam (2011) DNA barcoding as a means for identifying medicinal plants of Pakistan. Pakistan Journal of Botany 431-434.
- Chase MW, Fay MF (2009) Ecology. barcoding of plants and fungi. Science 325(5941): 682-683.
- Antil S, Abraham JS, Sripoorna D, Maurya S, Dagar J (2023) DNA barcoding, an effective tool for species identification: A review. Molecular biology reports 50(1): 761-775.
- Hubert F, Grimm GW, Jousselin E, Berry V, Franc A (2014) Multiple nuclear genes stabilize the phylogenetic backbone of the genus Quercus. Systematics and Biodiversity 12(4): 405-423.
- Sheth BP, Thaker VS (2017) DNA barcoding and traditional taxonomy: An integrated approach for biodiversity conservation. Genome 60(7): 618-628.
- Sajjad S, Islam M, Muhammad K, Ghafoor SU, Ullah I (2024) Comprehensive evaluation of cryptic juglans genotypes: Insight from molecular markers and phylogenetic analysis. Genes 15(11): 1417.
- Bolson M, Smidt EC, Brotto ML, Silva VP (2015) ITS and trnH-psbA as efficient DNA barcodes to identify threatened commercial woody angiosperms from southern Brazilian Atlantic rainforests. PloS one 10(12): e0143049.
- Ahmed SS (2022) DNA barcoding in plants and animals: A critical review.
- Dong W, Cheng T, Li C. Xu P, Long P, et al. (2014) Discriminating plants using the DNA barcode rbc L b: An appraisal based on a large data set. Molecular Ecology Resources 14(2): 336-343.
- Saddhe AA, Kumar K (2018) DNA barcoding of plants: Selection of core markers for taxonomic groups. Plant Science Today 5(1): 9-13.
- Schenk JJ, Becklund LL, Carey SJ, Fabre PP (2023) What is the “modified” CTAB protocol? Characterizing modifications to the CTAB DNA extraction protocol. Applications in Plant Sciences 11(3): 11517.
- Hollingsworth LL, Forrest LL, Spouge JL, Hajibabaei M (2009) A DNA barcode for land plants. Proc Natl Acad Sci USA 106(31): 12794-12797.
- Li KB (2003) ClustalW-MPI: ClustalW analysis using distributed and parallel computing. Bioinformatics 19(12): 1585-1586.
- Pang X, Liu H, Wu S, Yuan Y, Li H (2019) Species identification of oaks (Quercus, Fagaceae) from gene to genome. International Journal of Molecular Sciences 20(23): 5940.
- Yang J, Vázquez L, Chen X, Li H, Zhang H (2017) Development of chloroplast and nuclear DNA markers for Chinese oaks (Quercus subgenus Quercus) and assessment of their utility as DNA barcodes. Frontiers in Plant Science 8: 816.
- Blanc JC, Liesebach M (2015) Tracing the origin and species identity of Quercus robur and Quercus petraea in Europe: A review. Silvae Genetica 64(1-6): 182-193.
- Rellstab C, Zoller S, Walthert L, Lesur I, Pluess AR (2016) Signatures of local adaptation in candidate genes of oaks (Quercus) with respect to present and future climatic conditions. Molecular Ecology 25(23): 5907-5924.
- Sork VL, Fitz GST, Puiu D, Crepeau M, Gugger PF (2016) First draft assembly and annotation of the genome of a California endemic oak Quercus lobata Née (Fagaceae). G3 (Bethesda) 6(11): 3485-3495.
- Dodd RS, Afzal RZ (2004) Selection and dispersal in a multispecies oak hybrid zone. Evolution 58(2): 261-269.
- Li X, Li Y, Kassaby EY, Fang Y (2024) Diverse leaf functional and phenotypic traits in hybrid oak individuals: Unveiling genetic variation, adaptation, and evolutionary potential within Quercus Forest Ecology and Management 561: 121830.
- Curtu AL, Gailing O, Finkeldey R (2007) Evidence for hybridization and introgression within a species-rich oak (Quercus) community. BMC Evolutionary Biology 71-15.
- Fortini P, Pietro RD, Proietti E, Cardoni S, Quaranta L (2024) Dissecting the continuum and unravelling the phylogeographic knot of plastid DNA in European white oaks (Quercus Quercus): Ancient signatures and multiple diversity reservoirs. European Journal of Forest Research 143(1): 107-127.
- Fu R, Zhu Y, Liu Y, Feng Y, Lu RS (2022) Genome-wide analyses of introgression between two sympatric Asian oak species. Nature Ecology & Evolution 6(7): 924-935.
- Ginwal HS, Rawat A, Shekhar C, Bhandari MS, Kavidayal H (2024) Population genetic structure of a timberline oak (Quercus semecarpifolia ) of western Himalayas and conservation implications. Conservation Genetics 25(1): 133-147.
- Eaton EG, Caudullo S, Oliveira, De Rigo D (2016) Quercus robur and Quercus petraea in Europe: Distribution, habitat, usage and threats. European atlas of forest tree species pp: 160-163.
- Guirao RSS, Gonzalez J (2021) Benchmarking the performance of Pool‐seq SNP callers using simulated and real sequencing data. Molecular Ecology Resources 21(4): 1216-1229.
- Mc Shea WJ, Healy WM, Devers P, Fearer T, Koch FH (2007) Forestry matters: Decline of oaks will impact wildlife in hardwood forests. The Journal of Wildlife Management 71(5): 1717-1728.
- Patwardhan A, Ray S, Roy A (2014) Molecular markers in phylogenetic studies-a review. Journal of Phylogenetics & Evolutionary Biology 2(2): 131.
- Simpson MG (2019) Plant systematics. Academic press, USA.
- Hubert N, Hanner R (2015) DNA barcoding, species delineation and taxonomy: A historical perspective. DNA barcodes 3(1).
- Bidyananda N, Jamir I, Nowakowska K, Varte V, Vendrame WA (2024) Plant genetic diversity studies: Insights from DNA marker analyses. International Journal of Plant Biology 15(3): 607-640.
- Yang Y, Zhou T, Qian Z, Zhao G (2021) Phylogenetic relationships in Chinese oaks (Fagaceae, Quercus): Evidence from plastid genome using low-coverage whole genome sequencing. Genomics 113(3): 1438-1447.
- Wolfender JL, Nuzillard JM, Hooft JJ, Renault JH, Bertrand S (2018) Accelerating metabolite identification in natural product research: Toward an ideal combination of liquid chromatography–high-resolution tandem mass spectrometry and NMR profiling, in silico databases, and chemometrics. Analytical Chemistry 91(1): 704-742.
- Heise W, Babik W, Kubisz D, Kajtoch L (2015) A three-marker DNA barcoding approach for ecological studies of xerothermic plants and herbivorous insects from central Europe. Botanical journal of the Linnean Society. 177(4): 576-592.
- Cavender BA, Gonzalez RA, Pahlich A, Koehler K, Deacon N (2011) Phylogeography and climatic niche evolution in live oaks (Quercus series Virentes) from the tropics to the temperate zone. Journal of Biogeography 38(5): 962-981.
- Mc Vay JD, Hipp AL, Manos PS (2017) A genetic legacy of introgression confounds phylogeny and biogeography in oaks. Proc Biol Sci 284(1854): 20170300.
- Negi G, Samal P, Kuniyal J, Kothyari B, Sharma R (2012) Impact of climate change on the western Himalayan Mountain ecosystems: An overview. Tropical Ecology 53(3): 345-356.
- Potter KM, Jetton RM, Bower A, Jacobs DF. Man G (2017) Banking on the future: Progress, challenges and opportunities for the genetic conservation of forest trees. New Forests 48: 153-180.
- Awad M, Fahmy RM, Mosa KA, Helmy M. Feky FA (2017) Identification of effective DNA barcodes for Triticum plants through chloroplast genome-wide analysis. Computational Biology and Chemistry 71: 20-31.
- Ford CS, Ayres KL, Toomey N, Haider N, Alphen JVS (2009) Selection of candidate coding DNA barcoding regions for use on land plants. Botanical Journal of the Linnean Society 159(1): 1-11.
- Jiang S, Chen F, Qin P, Xie H, Peng G (2022) The specific DNA barcodes based on chloroplast genes for species identification of Theaceae Physiol Mol Biol Plants 28(4): 837-848.
- Kress WJ (2017) Plant DNA barcodes: Applications today and in the future. Journal of Systematics and Evolution 55(4): 291-307.
- Kress WJ, Erickson DL (2008) DNA barcoding: A windfall for tropical biology? Biotropica 40(4): 405-408.
- Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH (2005) Use of DNA barcodes to identify flowering plants. Proceedings of the National Academy of Sciences 102(23): 8369-8374.
© 2025 Abdul M. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






