- Submissions

Full Text
Approaches in Poultry, Dairy & Veterinary Sciences
Rumen Methanogens Diversity Analysis in Indian Buffaloes Using PCR-Denaturing Gradient Gel Electrophoresis
Malik PK*, Kolte AP, Bakshi B, Trivedi S and Bhatta R
ICAR-National Institute of Animal Nutrition and Physiology, Bengaluru, India
*Corresponding author: Malik PK, ICAR- National Institute of Animal Nutrition and Physiology, Bengaluru, India
Submission: June 19, 2020;Published: July 23, 2020

ISSN: 2576-9162 Volume7 Issue5
Abstract
A study aiming to investigate the rumen methanogens diversity using PCR- DGGE in Indian buffaloes (N= 10) was undertaken. Rumen fluid samples were collected from the male fistulated buffaloes. DNA from the rumen fluid samples was isolated using RBB+C method. DGGE gel band pattern was analysed and bands with differential expression were excised, cloned and sequenced for exploring the methanogens diversity in buffaloes. Sequences were aligned using Codon Code Aligner. The phylogeny was constructed based on neighbour joining method with Jukes Cantor nucleotide substitution model. Based on the differential patterns, total 31 bands were analyzed for the methanogen diversity through phylogenetic approach. Results from the study established that Methanobrevibacter is the most prominent genus of methanogens in Indian buffaloes. However, a considerable diversity exists within this genus at species level. Methanobrevibacter woesi represented the largest cluster of methanogens; while Methanobrevibacter millerae constituted the second largest fraction of archaeal community. Methanogens affiliated to Methanomicrobiales and Thermoplasmatales were not detected through DGGE in present study. To identify the methanogens from other minor groups such as Methanomicrobiales, Thermoplasmatales, Methanosarcinales further studies using high throughput techniques are warranted in buffaloes.
Keywords: Archaea; Buffalo; DGGE; Methanogens; Rumen
Introduction
Buffaloes with a global population of 206.6 millions remain an important ruminant species throughout the world [1]. India harbours 109.85 million buffaloes [2], which is about 53% of the global buffaloes population. Although, buffaloes are one of the important sources of animal origin food in the country, nevertheless enteric methane emission from them always remains an important concern. In addition to the contribution in global warming, enteric methane from the ruminants also epitomizes a significant loss of feed energy [3]. The annual enteric methane emission from the global buffaloes is about 11.05 Tg (teragram), whereas Indian buffaloes contribute 2.91 Tg [4]. Methanogens are unicellular organism belong to the domain Archaea and phylum Euryarchaeota [5]. Methane is a metabolic by-product produced by methanogens in strictly anaerobic conditions. Till date, 155 methanogens species belonging to 29 genera, 14 families and 6 orders have been isolated from the different ecosystems [6]. Methanogens are present in the rumen between an abundance of 107-1010 per gram of rumen content [7]. The substrate requirement of rumen methanogens is diverse, and majority of the archaea utilize carbon dioxide and hydrogen, while some of them prefers methanol, and methylamines [8] or formate [9] as substrate. Isolation of the archaea from rumen always remains a tedious task and that is why only limited (~10) species have been isolated till now [10,11]. Due to the culturing difficulties, it is likely that major fraction of the rumen methanogens is still to be identified [12,13]. Understanding of the rumen archaea is of utmost importance to develop the effective enteric methane mitigation strategies.
In spite of the better capabilities to adapt harsh conditions, high productivity and feed conversion efficiency, worldwide the studies in water buffaloes revealing methanogens di- versity are scanty. Limited studies have been conducted in Indian buffaloes [14-20], however, findings from these studies remained controversial. For example, most of these studies have reported Methanomicrobiales affiliated methanogens as most dominant in the buffalo rumen. However, global studies in buffaloes [21,22] and other species [23,24] concluded the presence of Methanomicrobiales methanogens to a limited extent in the rumen. Considering the inconclusive findings, small sampling size (3-4), and importance of diversity analysis for devising effective methane mitigation strategies, this study was undertaken to explore the rumen methanogens diversity in buffaloes through PCR based Denaturing Gradient Gel Electrophoresis (DGGE). In current study, we hypothesized that the rumen archaeal community is highly diverse and different methanogens species are distributed at a variable frequency.
Material and Method
Rumen liquor samples were collected from the buffaloes (N=10) to explore rumen methanogens diversity. Male fistulated buffaloes used as donor for the rumen liquor samples in this study. Animals were fed on a mixed diet comprising straw and concentrate in the proportion of 70:30. The necessary approval for collection of samples was obtained from the Institute Animal Ethics Committee (25/8/2016-CPCSEA part-I). Approximately, 50ml of digesta including solid and liquid fractions were collected from each animal through fistula and squeezed through double layers of muslin cloths. Aliquots of 15ml of the filtrate were transferred into sterile Eppendorf tubes and placed on the dry ice for transportation to the laboratory. The samples were stored in freezer at -20 °C till processed for the DNA isolation.
DNA Isolation and amplificationDNA from the rumen contents was extracted as per the RBB+C method of Yu and Morrison [25] using a Mini-Bead beater (Biospec, USA) plus column filtration with QIAamp DNA Mini Kit following the manufacturer’s instructions (Qiagen, GmbH). The microbial DNA was quantified with bio-spectrometer (Eppendorf, Germany) and assessed for the quality on 1.2% agarose gel. To explore the rumen methanogens diversity, a PCR (50μl reaction) was performed with DGGE specific primers Arch 344F (GC clamp) ‘GCC GCC CGC CGC GCG CGG CGG GCG GGG CGG GGG CAC GGG GGG ACG GGG ACG AGC AGG CGC GA’ and 522R ‘GWA TTA CCG CGG CKG CTG 3’ to amplify the variable region of the 16S rDNA [26]. Following conditions were maintained during the PCR amplification: initial denaturation 94 ̊C 3min; 94 ̊C 30s, 65 ̊C 30s, 72 ̊C 30s (20 cycles); 94 ̊C 30s, 55 ̊C 30s, 72 ̊C 30s (15 cycles); 72 ̊C 10min followed by holding of amplified product at 4 ̊C.
DGGE and sequencingA 8% polyacrylamide gel was prepared with a denaturant gradient between 30% and 60% urea and formamide. About 30μlamplified sample was loaded in each well and electrophoresed for 18hrs at 60 ̊C using a fixed voltage of 80V. After termination of electrophoresis, the gel was stained with silver solution to visualize the bands. The banding pattern was auto detected and the dendrogram was generated for the lane profiles of the DGGE gel in GelQuest and ClustVis software (SequentiX, Germany). An UPGMA tree was constructed with unweighted pair group method using arithmetic average distance, Dice and bootstraps. The tree was visualized on https://itol.embl.de. The bands were excised using a clean, sharp scalpel and placed into a 1.5ml tube. DNA was eluted from polyacrylamide gel slices according to the method of [27]. Another PCR was setup using eluted samples as template (2μl); however, forward primer i.e. 344F without GC clamp ‘ACG GGG CGC AGC AGG CGC GA’ was used this time; while reverse primer remained the same. The amplification was confirmed by running PCR product on 1.2% agarose gel. Libraries of 16S rDNA of total archea were prepared from PCR product through ligation (pJET vector) and transformation into Escherichia coli KRX competent cell. The amplicons were sent for the sequencing using Sanger chemistry (ABI 3730x1, Applied Biosystems). Sequences were aligned using Codon Code Aligner (version V4.0.4), removed the primer sequences and edited manually maintaining the alignment score above Q20. The sequences from the DGGE bands were aligned along with the references archaeal sequences downloaded from Ribosomal Database Project on CLC Genomics Workbench (v 20.0, Qiagen, Germany). The phylogeny was constructed based on neighbour joining method with Jukes Cantor nucleotide substitution model with 1000 bootstraps. The resultant tree was visualized and annotated on https://itol.embl. de.
Results
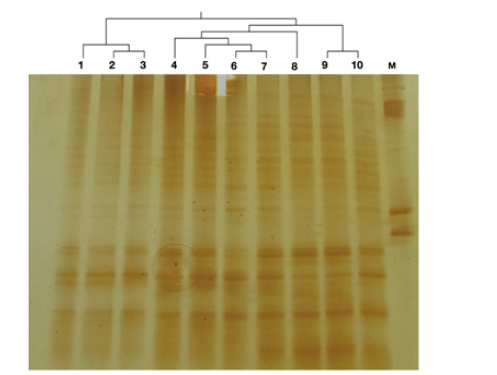
Primer pair Arch 344F/522R generated clear PCR products from rumen archaeal samples but did not amplify the bacterial DNA. Amplicons of approximately 200bp lengths were obtained from the amplification of archaeal DNA and no band from the unwanted DNA fragment (non-specific) was observed through visualization on 1.2% agarose gel. In DGGE, DNA strands were separated based on their actual base composition or GC: AT ratio. PCR fragments generated with selected primer pair were analyzed by DGGE (Figure 1). Band patterns with DGGE showed good resolution and separation pattern among the samples. Analysis of DGGE gel showed significant variation in the band patterns among buffalo samples, which revealed the difference in the rumen archaeal community. Rumen samples 1, 2 and 3 were similar in the band pattern and rumen archaeal distribution; while samples 4, 5, 6 and 7 were in close relatedness with sample 8. Sample 9 and 10 were apart from the above two cluster and grouped together as far as band pattern is concerned.
Based on the differential patterns, a total 31 bands were excised and reamplified using the same primer pair without GC clamp. Sequences from the excised bands were analyzed for the methanogen diversity through phylogenetic approach. All fragments belonged to archaea and shown the similarity with the methanogens from Methanobrevibacter genus. However, a considerable diversity was noted in the archaeal distribution at species level within this genus. The fragments were clustered into eight groups. Largest cluster constituting nine of the 31 fragments have shown the similarity with Methanobrevibacter woesi (Figure 2); while second largest cluster with seven fragments have shown the maximum similarity with Methanobrevibacter millerae. Four fragments were separated and clustered with the Methanobrevibacter gottschalkii. Similarly, three fragments have shown the identity similar to Methanobrevibacter oralis. In present study, six fragments (two of each) were similar to the Methanobrevibacter thaueri, Methanobrevibacter smithii and Methanobrevibacter boviskoreani. This study established that the methanogens from Methaobrevibacter genus with highest representation of Methanobrevibacter woesi and Methanobrevibacter milleare were highly abundant in the buffalo rumen. All the methanogens in present study belonged to the hydrogenotrophic category of revealing that methanogenesis through hydrogenotrophic pathway is the most prevalent in Indian buffaloes. Methanogens from the RO clade within Methanobrevibacter genus, Methanomicrobiales and Thermoplasmatales were not identified in this study.
Figure 1: The 16S ribosomal DNA denaturing gradient gel electrophoresis (DGGE) of Buffalo rumen archaeal community fingerprint analysis. The samples are clustered using UPGMA dendrogram on the lane profiles.

Figure 2: Phylogenetic tree of partial 16S rRNA gene sequences of DGGE bands obtained from different buffalo rumen samples. Reference strain sequences are downloaded from the RDP database and their respective Genbank accession numbers are given in the parenthesis.
Discussion
Methanogens are vital in driving electro flow in the rumen and sustaining the metabolism. Archaea are majorly involved in the reduction of carbon molecules with various electron donors [28]. Methanogens constitute 2-4% of the total microbes in rumen [29,30] and their presence in the rumen has been reported from 20 minutes [31] to few days of the birth [32,33]. About 10 methanogens have been isolated from the rumen till date; however, 16s rRNA based analysis confirmed the presence of a lot more methanogens in the rumen [24]. Thus, a larger fraction of the archaeal community yet to be elucidated. In general, buffaloes have better feed conversion efficiency than cattle and efficient producer. They are best suited to the prevailing feeding practices and harsh conditions in the country. A significant variation in the rumen microbial population had been reported between cattle and buffaloes [34,35]. Globally, the rumen microbiota in general and methanogens community in particular is less characterized as compared to cattle. Though the rumen archaeal community is less diverse than bacteria, nevertheless, many factors such as host species and geographical location can bring the difference in community structure [24]. Archaeal community composition rather total intensity have more impact on the methane emission [36]. For example, Methanobrevibacter SGMT clade has great influence on the methane emission than any other clade of methanogens [37]. Efficient utilization capacity of H2 by SGMT clade methanogens has reported one of the major cause for their great influence on methane emission [38].
In this study, archaea belonging to the genus Methanobrevibacter were reported; however, 31 fragments were clustered into seven species. Two largest cluser of the fragments were similar to the Methanobrevibacter woesi and Methanobrevibacter millerae (Figure 2). Methanogens affiliated to Mb. gottschalkii, Mb. thaueri, Mb. smithii, Mb. oralis and Mb. boviskorenai were also identified in the buffaloes rumen through DGGE. In another study by our group, the methanogens affiliated to the Methanobrevibacter genus were reported most abundant constituted 98.4% of the clone libraries from buffalo rumen fluid samples (Malik et al., communicated). The results of our study concurs with the previous reports [10,39,40]. Results from our study are in good agreement with the Franzolin R et al. [21] and Iqbal MWet al. [22], reported Methanobrevibacter as most dominant genus. Methanogens from this genus majorly falls into two groups i.e. SGMT and RO clade. Our study identified the methanogens from SGMT clade in buffaloes rumen; while methanogens affiliated to the RO clade remained unidentified. There are many global datasets identified the methanogens from these two clades in different ruminant species [41-44]; however, a remarkable variation in the abundance at species level was noticed. Similar to present study, Methanobrevibacter boviskoreani was also identified in the Korean native cattle but at a low frequency [11]. The disagreement for the methanogens abundance at species level between this study and previous reports could be attributed to the host species, diet, geographical region, DNA isolation and downstream processing. In a recent cloning based study, the impact of host species (cattle & buffalo) on methanogens diversity was explored (Malik et al., communicated). They concluded that Methanobrevibacter millerae was most dominating species of methanogens in Indian cattle; while, Methanobrevibacter smithii occupied the major niche of archaeal community in buffaloes (Malik et al., communicated). Wright ADG et al. [44,45] from their studies concluded that the geographical location has an impact on rumen methanogens community composition.
Contrary to our findings, the previous Indian reports revealed Methanomicrobiales as the most abundant methanogens in Indian buffaloes [14,16]. However, in another comprehensive study, Methanomicrobiales abundance was found less than 5% of the total archaeal distribution in Indian buffaloes (Malik et al., communicated). Similarly, global studies have also not confirmed the Methanomicrobiales as dominant methanogens in ruminants [23,24]. Methanobacterium were not identified in buffaloes; however, surprisingly their distribution has been reported up to 63% in Nili-Ravi buffalo by Paul SS et al. [19]. Methanogens affiliated to Methanosphaera, RCC clade, Methanosarcina were also not identified in buffaloes through DGGE. It has been reported that RCC clade methanogens may not be universally present in livestock [42,46,47]. Our results are in agreement with Jeyanathan J et al. [24] who also have not reported the RCC group methanogens in a DGGE study. Detection of RCC sequences in DGGE patterns of the archaeal 16S rRNA genes remained extremely difficult, which could be reason for the absence of methanogens from this group in our study. Although the PCR-DGGE technique is very useful in identifying the microbial diversity [48,49] among the samples, nevertheless the detection always remained restricted to prominent species [50]. Each band on the DGGE gel should affiliate to one sequence/species of the microbe; however, multiple bands in our study were found representing one species. This is not surprising and has been reported previously [51]. Total 31 fragments in present study represented seven species of methanogens all affiliated to the Methanobrevibacter. The limited fragment length in DGGE limits the sequence information for phylogenetic analysis [52]. The possible explanation for the non-detection of other rumen methanogens in Indian buffaloes could be their limited representation in the overall rumen archaeal community. It is obvious that DNA samples can be separated into thousands different fragments, whereas only 30-40 fragments can be visualized in DGGE [50]. Therefore, the archaea with limited abundance in the rumen usually remained undetected in the DGGE.
Conclusion
From the study, it can be concluded that Methanobrevibacter is the most dominating genus of methanogens in Indian buffaloes. This genus is highly diversified and Methanobrevibacter woesi represented the largest cluster of methanogens in the buffalo rumen; while Methanobrevibacter millerae constituted the second largest niche of the archaeal community. Other important species of methanogens such as Methanobrevibacter gottschalkii, Methanobrevibacter oralis, Methanobrevibacter thaueri, Methanobrevibacter smithii and Methanobrevibacter boviskoreani were also detected in the buffalo rumen. However, methanogens with limited representation in the rumen archaeal community and affiliated to Methanomicrobiales and Thermoplasmatales were not detected through DGGE in present study. Hydrogenotrophic methanogens were prevalent in the buffalo rumen. To identify the methanogens from other minor groups such as Methanomicrobiales, Thermoplasmatales, Methanosarcinales further studies using high throughput techniques are warranted in buffaloes.
Acknowledgement
The authors are thankful to the Department of Biotechnology (DBT), New Delhi for extending the financial support to carry out this research under a project entitled “Livestock Methane Reduction through Immunization based Approach (BT/PR8750/AAQ/1- 555/2013)”. The authors also wish to thank the Director of the institute for his kind support in completing the project.
Conflict of Interest
The authors report no conflicts of interest.
References
- (2020) FAOSTAT, Rome, Italy.
- (2019) 20th Livest census provisional key results. Department of Animal Husbandry and Dairying, Government of India.
- Czerkawski JW (1969) Methane production in ruminants and its significance. World Rev Nutr Diet 11: 240-282.
- Bhatta R, Malik PK, Kolte AP, Suresh KP (2009) Assessment of enteric methane emission from Indian livestock: A new approach. In: Sejian V, Isloor S, Rahman SA, Bhatta R (Eds.), 7th Pan Commonw Vet Conf Commonwealth Veterinary Association (Asia), Bengaluru, pp. 101-103.
- Woese CR, Kandler O, Wheelis ML (1990) Towards a natural system of organisms: Proposal for the domains archaea, bacteria, and eucarya. Proc Natl Acad Sci USA 87(12): 4576-4579.
- Holmes DE, Smith JA (2016) Biologically Produced Methane as a Renewable Energy Source. Adv Appl Microbiol, Elsevier, Netherlands, 97: 1-61.
- Joblin KN (2005) Methanogenic archaea. In: MHP, MCS (Eds.), Methods Gut Microb Eco Ruminants, pp. 47-53.
- Patterson JA, Hespell RB (1979) Trimethylamine and methylamine as growth substrates for rumen bacteria and Methanosarcina barkeri. Curr Microbiol 3: 79-83.
- Hungate RE, Smith W, Bauchop T, Yu I, Rabinowitz JC (1970) Formate as an intermediate in the bovine rumen fermentation. J Bacteriol 102(2): 389-397.
- Janssen PH, Kirs M (2008) Structure of the archaeal community of the rumen. Appl Environ Microbiol 74(12): 3619-3625.
- Lee JH, Kumar S, Lee GH, Chang DH, Rhee MS, et al. (2013) Methanobrevibacter boviskoreani sp. nov., isolated from the rumen of Korean native cattle. Int J Syst Evol Microbiol 63(Pt 11): 4196-4201.
- Raskin L, Stromley JM, Rittmann BE, Stahl DA (1994) Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol 60(4): 1232-1240.
- Wolin MJ, Miller TL, Stewart CS (1997) Microbe-microbe interactions. In: Hobson PN, Stewart CS (Eds.), Rumen Microb Ecosyst, Springer Netherlands, Dordrecht pp. 467-491.
- Singh KM, Tripathi AK, Pandya PR, Parnerkar S, Rank DN, et al. (2012) Methanogen diversity in the rumen of Indian Surti buffalo (Bubalus bubalis), assessed by 16S rDNA analysis. Res Vet Sci 92(3): 451-455.
- Singh KM, Tripathi AK, Pandya PR, Parnerkar S, Kothari RK et al. (2013) Molecular genetic diversity and quantitation of methanogen in ruminal fluid of buffalo (Bubalus bubalis) fed ration (Wheat straw and concentrate mixture diet). Genet Res Int 2013: 1-7.
- Chaudhary PP, Sirohi SK (2009) Dominance of Methanomicrobium phylotype in methanogen population present in Murrah buffaloes (Bubalus bubalis). Lett Appl Microbiol 49(2): 274-277.
- Parmar NR, Nirmal Kumar JI, Joshi CG (2015) Exploring diet-dependent shifts in methanogen and methanotroph diversity in the rumen of Mehsani buffalo by a metagenomics approach. Front Life Sci 8(4): 371-378.
- Kumar S, Dagar SS, Puniya AK (2012) Isolation and characterization of methanogens from rumen of Murrah buffalo. Ann Microbiol 62: 345-350.
- Paul SS, Deb SM, Dey A, Somvanshi SPS, Singh D, et al. (2015) 16S rDNA analysis of archaea indicates dominance of Methanobacterium and high abundance of Methanomassiliicoccaceae in rumen of Nili-Ravi buffalo. Anaerobe 35(Pt B): 3-10.
- Chaudhary PP, Sirohi SK, Saxena J (2012) Diversity analysis of methanogens in rumen of Bubalus bubalis by 16S riboprinting and sequence analysis. Gene 493(1): 13-17.
- Franzolin R, Pierre BS, Northwood K, Wright ADG (2012) Analysis of rumen methanogen diversity in water buffaloes (Bubalus bubalis) under three different diets. Microb Ecol 64(1): 131-139.
- Iqbal MW, Zhang Q, Yang Y, Li L, Zou C, et al. (2018) Comparative study of rumen fermentation and microbial community differences between water buffalo and jersey cows under similar feeding conditions. J Appl Anim Res 46(1): 740-748.
- Henderson G, Cox F, Ganesh S, Jonker A, Young W, et al. (2015) Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci Rep 5: 14567.
- Jeyanathan J, Kirs M, Ronimus RS, Hoskin SO, Janssen PH (2011) Methanogen community structure in the rumens of farmed sheep, cattle and red deer fed different diets. FEMS Microbiol Ecol 76(2): 311-326.
- Yu Z, Morrison M (2004) Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 36(5): 808-812.
- Chan OC, Wolf M, Hepperle D, Casper P (2002) Methanogenic archaeal community in the sediment of an artificially partitioned acidic bog lake. FEMS Microbiol Ecol 42(1): 119-129.
- Etokebe GE, Spurkland A (2000) Method for avoiding PCR-inhibiting contaminants when eluting DNA from polyacrylamide gels. Biotechniques 29(4): 694-696.
- Friedman N, Shriker E, Gold B, Durman T, Zarecki R, et al. (2017) Diet‐induced changes of redox potential underlie compositional shifts in the rumen archaeal community. Environ Microbiol 19(1): 174-184.
- Xue D, Chen H, Chen F, He Y, Zhao C, et al. (2016) Analysis of the rumen bacteria and methanogenic archaea of yak (Bos grunniens) steers grazing on the Qinghai-Tibetan Plateau. Livest Sci 188: 61-71.
- Zhou M, McAllister TA, Guan LL (2011) Molecular identification of rumen methanogens: Technologies, advances and prospects. Anim Feed Sci Technol 166-167: 76-86.
- Guzman CE, Bereza-Malcolm LT, De Groef B, Franks AE (2015) Presence of selected methanogens, fibrolytic bacteria, and proteobacteria in the gastrointestinal tract of neonatal dairy calves from birth to 72 hours. Plos One 10(7): e0133048.
- Morvan B, Dore J, Lesme FR, Foucat L, Fonty G, et al. (1994) Establishment of hydrogen-utilizing bacteria in the rumen of the newborn lamb. FEMS Microbiol Lett 117(3): 249-256.
- Skillman LC, Evans PN, Naylor GE, Morvan B, Jarvis GN, et al. (2004) 16S ribosomal DNA-directed PCR primers for ruminal methanogens and identification of methanogens colonising young lambs. Anaerobe 10(5): 277-285.
- Wanapat M, Chanthakhoun V, Wanapat M, Chanthakhoun V (2009) Recent advances in rumen ecology, digestion and feeding strategies of swamp buffaloes. Simpósio Búfalos Das Américas 5: 27-36.
- Calabrò S, Moniello G, Piccolo V, Bovera F, Infascelli F, et al. (2008) Rumen fermentation and degradability in buffalo and cattle using the in vitro gas production technique. J Anim Physiol Anim Nutr (Berl) 92: 356-362.
- Tapio, Snelling TJ, Strozzi F, Wallace RJ (2017) The ruminal microbiome associated with methane emissions from ruminant livestock. J Anim Sci Biotechnol 8: 7.
- Danielsson R, Dicksved J, Sun L, Gonda H, Müller B, et al. (2017) Methane production in dairy cows correlates with rumen methanogenic and bacterial community structure. Front Microbiol 8: 226.
- Mcallister TA, Meale SJ, Valle E, Guan LL, Zhou M, et al. (2015) Ruminant nutrition symposium: Use of genomics and transcriptomics to identify strategies to lower ruminal methanogenesis. J Anim Sci 93: 1431-1449.
- Miller TL (1994) Ecology of methane production and hydrogen sinks in the rumen. Proc Soc Nutr Physiol, DLG, Germany.
- Wright ADG, Auckland CH, Lynn DH (2007) Molecular diversity of methanogens in feedlot cattle from Ontario and Prince Edward Island, Canada. Appl Environ Microbiol 73(13): 4206-4210.
- Shin EC, Choi BR, Lim WJ, Hong SY, An CL, et al. (2004) Phylogenetic analysis of archaea in three fractions of cow rumen based on the 16S rDNA sequence. Anaerobe 10(6): 313-319.
- Skillman LC, Evans PN, Strömpl C, Joblin KN (2006)16S rDNA directed PCR primers and detection of methanogens in the bovine rumen. Lett Appl Microbiol 42(3): 222-228.
- Whitford MF, Teather RM, Forster RJ (2001) Phylogenetic analysis of methanogens from the bovine rumen. BMC Microbiol 1: 1-5.
- Wright ADG, Williams AJ, Winder B, Christophersen CT, Rodgers SL, et al (2004) Molecular diversity of rumen methanogens from sheep in western Australia molecular diversity of rumen methanogens from sheep in western Australia. Appl Environ Microbiol 70(3): 1263-1270.
- Wright ADG, Toovey AF, Pimm CL (2006) Molecular identification of methanogenic archaea from sheep in Queensland, Australia reveal more uncultured novel archaea. Anaerobe 12(3): 134-139.
- Wright ADG, Ma X, Obispo NE (2006) Methanobrevibacter phylotypes are the dominant methanogens in sheep from Venezuela. Microb Ecol 56(2): 390-394.
- Zhou M, Sanabria EH, Le LG (2009) Assessment of the microbial ecology of ruminal methanogens in cattle with different feed efficiencies. Appl Environ Microbiol 75(20): 6524-6533.
- Kocherginskaya SA, Aminov RI, White BA (2001) Analysis of the rumen bacterial diversity under two different diet conditions using denaturing gradient gel electrophoresis, random sequencing, and statistical ecology approaches. Anaerobe 7(3): 119-134.
- Regensbogenova M, Kisidayova S, Michalowski T, Javorsky P, Van Der Staay SYM, et al, (2004) Rapid identification of rumen protozoa by restriction analysis of amplified 18S rRNA gene. Acta Protozool 43: 219-224.
- Christophersen C (2007) Grain and artificial stimulation of the rumen change the abundance and diversity of methanogens and their association with ciliates, University of Western Australia, Australia.
- Yu Z, González RG, Schanbacher FL, Morrison M (2008) Evaluations of different hypervariable regions of archaeal 16S rRNA genes in profiling of methanogens by Archaea-specific PCR and denaturing gradient gel electrophoresis. Appl Environ Microbiol 74(3): 889-893.
- Myers RM, Fischer SG, Lerman LS, Maniatis T (1985) Nearly all single base substitutions in DNA fragments joined to a GC-clamp can be detected by denaturing gradient gel electrophoresis. Nucleic Acids Res 13(9): 3131-3145.
© 2020 Malik PK. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






