- Submissions

Full Text
Aspects in Mining & Mineral Science
Optimisation of a Sustainable Method for DNA Extraction in Acidithiobacillus Ferrooxidans Using Non-Halogenated Buffers, for Improved Applications in Biomining Technology
Vidhya Sagar Murugesan*, Matthew Duffield Healy and Sebastien Farnaud
Research Centre for Health and Life Sciences, Coventry University, United Kingdom
*Corresponding author:Vidhya Sagar Murugesan, Research Centre for Health and Life Sciences, Coventry University, Whitefriars Street, Coventry CV1 5FB, United Kingdom
Submission: September 17, 2025: Published: September 30, 2025

ISSN 2578-0255Volume14 Issue 2
Abstract
DNA was extracted using the DNeasy PowerSoil Pro kit with TBE and PBS buffers, and the results were compared with those obtained using Tris-HCl and no buffer. After extraction, DNA concentration and purity were studied using a NanoDrop spectrophotometer. The DNA extracted using PBS had a higher concentration (220.23ng/μL), while the lowest concentration was observed in TBE (45.6ng/μL). The DNA concentration measured after PCR showed that the DNA extracted using the TBE 5mL buffer had the highest concentration (708ng/μL), while the lowest concentration was in PBS (125.48ng/μL). After PCR clean-up, the final DNA concentration was higher in PBS (244.5±10.26ng/μL) and TBE 10mL (56.87ng/μL) than in Tris-HCl (38.75ng/μL), with the lowest in 5mL TBE (13.85ng/μL). The DNA sample extracted without buffer showed the lowest yield (6.18ng/μL). Statistical analysis revealed significant differences between PBS and Tris-HCl (p=7.61×10⁻⁵), with PBS-extracted DNA exhibiting a higher concentration. These results show that both buffer composition and volume are critical for yield, and that non-halogenated buffers can be effective alternatives to Tris-HCl.
Keywords:DNA extraction; Acidithiobacillus ferrooxidans; TBE buffer; PBS buffer; Tris-HCl; Quantification; Non- halogenated buffers
Abbreviations:PBS: Phosphate-Buffered Saline; TBE: Tris-Borate-EDTA; PCR: Polymerase Chain Reaction
Introduction
The DNA extraction process is a critical stage in yield, safety, and applicability to the downstream process. The choice of buffer is, therefore, fundamental. At an earlier stage, conventional DNA extraction was performed using phenol-chloroform-isoamyl alcohol (25:24:1) [1], with Tris-HCl being a commonly used buffer in molecular biology, containing chloride ions (Cl⁻) to provide stability during rigorous purification [2]. These methods are generally efficient for DNA recovery. However, regarding environmental and health risks, they generate halogenated wastes, which are classified as group B2 carcinogens due to their potential to produce persistent compounds such as chloride and bromide that are difficult to degrade and environmentally harmful [3]. Consequently, researchers have explored more sustainable and less hazardous approaches to reduce byproduct formation. Recently, hydrophilic Ionic Liquids (ILs) have been introduced as novel extraction agents, capable of simultaneously lysing cells and solubilising DNA at high yield, although they can be more costly and require careful handling [4]. However, ILs represent the sustainable improvement of the conventional Tris-HCl and phenol-chloroform methods, driven by the increasing demand for environmentally friendly, efficient, and reliable DNA extraction strategies. Based on these advancements, the present study aims to optimise the DNA extraction procedure of Acidithiobacillus ferrooxidans increasingly used in Biomining, using non-halogenated buffers such as TBE and to compare their effectiveness, efficiency, and compatibility with downstream molecular techniques.
Methods
Bacterial culture and bacterial pre-treatment
The ABSTE medium was prepared by combining Acidophile Basal Salt (ABS) and Acidophile Trace Element (TE) in 100mL of deionised water (pH 1.8) and sterilised. In ABSTE medium, Iron(II) sulphate, elemental sulphur, and the isolated A. ferrooxidans strain were inoculated aseptically and cultures were incubated at 30 °C, 160 RPM in the New Brunswick Innova® Shaking incubator as previously described (Duffield-Healy & Farnaud, 2023). Bacterial culture was subjected to three trials: two with TBE buffer and one with PBS buffer. Triplicates of 25mL (TBE) and 15mL (PBS) cultures were centrifuged at 4000g for 10min (Fisherbrand™ GT2R, Thermo Fisher Scientific). The supernatant was discarded, and cell pellets were resuspended in 10×buffer (10mL for Trial 1, 5mL for Trial 2 in TBE; 10mL for Trial 3 in PBS) and vortexed. pH was confirmed to be above 4 for all samples. After centrifugation, the buffer was removed, 1mL MilliQ H₂O was added to form a slurry, transferred to a PowerSoil® Pro bead vial, centrifuged at 16000g for 1 min, and residual liquid was discarded.
DNA extraction and quantification
DNA of At. ferrooxidans was extracted using the DNeasy® PowerSoil® Pro manufacturer’s protocol and quantified using a Nanodrop spectrophotometry (NanoDrop™ One, Thermo Fisher Scientific). In the dsDNA option, 2μL of C6 solution was added. Then, 2μL of the extracted DNA was added.
DNA amplification using PCR and PCR clean up
Primers for At. ferrooxidans CYC1 were designed using NCBI Primer-BLAST based on the ATCC 23270 strain. Primers were reconstituted in nuclease-free water to prepare 100μM stocks and 10μM working solutions. PCR reactions (50μL) were prepared with template DNA, primers, and 5×master mix following standard protocols. Reactions were run on an Eppendorf Mastercycler® using the following conditions: initial denaturation at 95 °C for 30s, then the same conditions were repeated for 30 cycles. Then, it was run at 60°C for 1min, and 68 °C for 1min/kb. The final extension was at 68 °C for 5min and held at 4 °C. PCR clean-up was done using T1130S 50 preps Monarch® Spin PCR & DNA Cleanup Kit as described by the manufacturer’s protocol, and DNA was eluted using the Nuclease-free H2O.
Result and Discussion
The extraction of DNA and PCR amplification were performed using three different buffers: PBS, TBE 5ml, and TBE 10ml, as shown in Table 1. The major findings indicated significant variation in DNA concentration at the pre-PCR, post-PCR, and post-PCR cleanup stages, demonstrating that the choice of buffer played a crucial role in determining DNA concentration. As seen in Table 1, at the initial stage, the DNA extracted using PBS had a higher concentration (220.23ng/μL) than the TBE 5ml, 10ml, and Tris- HCl. During cell lysis, PBS might have played a role in maintaining pH while the DNA was released, resulting in DNA without shearing. Similar findings were reported by Fukushima et al. [5], who stated that the DNA rinsed in PBS maintained its double-stranded structure and its functional properties even after DNA-chitosan polymers were loosely packed, which demonstrated PBS’s role in DNA stability. The lowest value was for 5ml TBE, which indicated that the reduced volume of the TBE buffer affected cell lysis and led to a loss of efficiency in providing a good yield of DNA [6]. The post-PCR quantification showed a different result, in which TBE 10ml had the highest concentration of DNA (708ng/μL). The yield for PBS was only 125.48ng/μL; this may have been due to improper amplification. However, due to the presence of PCR inhibitors such as higher salt content and excess borate, which alter the ionic environment where magnesium ion complexes may have been formed [7]. This was confirmed by Nanodrop analysis, which showed peaks at A260/230 that denoted salt contamination [8].
Table 1:DNA concentration and relative yield (%) of Acidithiobacillus ferrooxidans at different stages using various buffers.
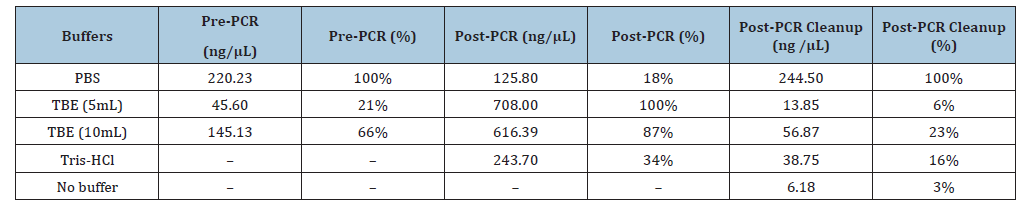
The results after the PCR cleanup showed that the PBS concentration was higher (244.5±10.26ng/μL) among all buffers. Whereas the DNA concentration of the TBE 5ml sample was reduced to 13.85ng/μL, and the lowest concentration was observed in the no-buffer sample (6.18ng/μL) as shown in Figure 1. The final concentration results showed that both the buffer and volume were essential for obtaining a good yield of DNA after PCR cleanup. The increase in buffer volume from 5ml to 10ml TBE enhanced DNA yield by improving cell lysis or reducing the impact of contaminants (QIAGEN, n.d.) [9]. PBS showed the same consistency pre- and post- PCR cleanup, where PBS maintained the pH and stability and hence minimised DNA loss and preserved the integrity. This aligned with the study on forensic swabs by Martin et al. [10], where PBS was reported to prevent spermatozoa DNA denaturation.
The Halogenated Tris-HCl buffer showed a lower yield (38.75±8.49ng/μL) compared to both non-halogenated buffers, (PBS and 10ml TBE buffer), as seen in Figure 1. This revealed that, PBS balanced the ionic strength also, the presence of EDTA and borate in the TBE buffer contributed to better retention of DNA during extraction and amplification. Environment-friendly buffers like TBE and PBS outperformed Tris-HCl, suggesting that they were better replacements for standard buffers. This was supported by OPS Diagnostics (n.d.) [11], which stated that even non-halogenated buffers like CTAB showed higher yield than commercial halogenated buffers like Tris-HCl. Furthermore, a fluorometric method like Qubit is recommended for precise DNA yield measurement [12], as the Nanodrop spectrophotometer has certain limitations. These limitations include low accuracy in detecting PCR inhibitors and the inability to distinguish between intact and denatured DNA. Future research should also consider testing other non-halogenated buffer systems and scaling the optimised conditions for high-throughput extractions in bioleaching applications.
Figure 1:DNA concentration (ng/μL) after PCR cleanup was analysed for TBE, PBS, Tris-HCl 5ml, 10ml and no buffer samples. DNA concentration (ng/μL) after PCR cleanup was analysed for TBE, PBS, Tris-HCl 5ml, 10ml and no buffer samples. PBS has the highest concentration of 244.5±10.26ng/μL, followed by TBE 10ml (56.88±6.3ng/μL), Tris- HCl (38.75±8.49ng/μL), TBE 5ml (13.9±5.35ng/μL) and no buffer (6.18±2.10ng/μL). Samples underwent statistical analysis by unpaired t-tests, which show significant differences between PBS and TBE 10ml (p=0.0001), PBS and Tris-HCl (p=7.61106×10-5), PBS and no buffer (p=9.48105E-05), PBS and TBE 5ml (p=7.66606E-05), Tris-HCl and no buffer (p=0.0276), TBE 10ml and no buffer (p=0.004), TBE 10ml vs TBE 5ml (p=0.01). All sample groups passed the Shapiro-Wilk normality test (p>0.05), approving parametric comparisons. Shapiro-Wilk test statistics are as follows: W value of TBE 5ml=1, TBE 10ml=0.9466, PBS=0.9357, Tris-HCl=0.9843, no buffer=0.7821; p value of TBE 5ml=1, TBE 10ml=0.915, PBS=0.74, Tris-HCl=0.99, no buffer=0.08. Significance was indicated as: *p<0.05, **p<0.01, ***p<0.001, ns=not significant.

Conclusion
As shown in the current study, the type and amount of buffer have significant effects on the efficiency of DNA extraction and the following PCR amplification. PBS demonstrated the ability to stabilise pH and retain the structural integrity of the DNA constituent throughout pre- and post-PCR, as well as pre- and post- cleanup, with the highest DNA yield. By contrast, TBE performance was sensitive to the volume of reaction; 10ml performed better than 5ml, presumably because all the cell lysis was converted to a positive effect, and the contamination effects were reduced. The use of non-halogenated buffers has proven to be beneficial since the DNA concentration was lower when tris-HCl, one of the halogenated buffers, was used. These findings support the theoretical assumption that buffer formulation is a relevant determinant of the number of DNA copies and the stability of DNA, and suggest a potential advantageous application of environmentfriendly and non-halogenated buffers, such as PBS or optimised volumes of TBE, to replace the theoretical assumption that buffer formulation is a relevant determinant of the number of copies of the DNA and the stability of DNA and find potential advantageous application of environment-friendly and non- halogenated buffers like PBS or optimised volumes of TBE to replace the traditional buffers that contain halogens. The findings confirm the supposition that the perfect choice of buffer plays a crucial role in maximising both the yield and integrity of DNA, with significant implications for its practical use in molecular as well as bioleaching applications.
References
- Sharma R, Sharad S, Minhas G, Sharma DR, Bhatia K, et al. (2023) DNA, RNA isolation, primer designing, sequence submission, and phylogenetic analysis. Basic Biotechniques for Bioprocess and Bioentrepreneurship pp. 197-206.
- Song Y, Fahs A, Feldman C, Shah S, Gu Y, et al. (2013) A reliable and effective method of DNA isolation from old human blood paper cards. SpringerPlus 2(1): 616.
- U S Environmental Protection Agency (2016) Chloroform (Hazard summary). Retrieved from U.S. Environmental Protection Agency website.
- Martzy R, Bica-Schröder K, Palvölgyi AM, Kolm C, Jakwerth S, et al. (2019) Simple lysis of bacterial cells for DNA-based diagnostics using hydrophilic ionic liquids. Scientific Reports 9(1): 13994.
- Fukushima T, Hayakawa T, Kawaguchi M, Ogura R, Inoue Y, et al. (2005) PBS buffer solutions with different pH values can change porosity of DNA-chitosan complexes. Dental Materials Journal 24(3): 414-421.
- Dilley K, Pagan F, Chapman B (2021) Methods for ensuring the highest DNA concentration and yield in future and retrospective trace DNA extracts. Science & Justice 61(2): 193-197.
- Sidstedt M, Ra˚dström P, Hedman J (2020) PCR inhibition in qPCR, dPCR and MPS-mechanisms and solutions. Analytical and Bioanalytical Chemistry 412(9): 2009-2023.
- Matlock B (2015) Assessment of nucleic acid purity (Technical Note 52646). Thermo Fisher Scientific.
- Qiagen (n.d.). Lysis of bacterial cells for plasmid purification.
- Martin NC, Pirie AA, Ford LV, Callaghan CL, McTurk K, et al. (2006) The use of phosphate buffered saline for the recovery of cells and spermatozoa from swabs. Science & Justice 46(3): 179-184.
- OPS Diagnostics (n.d.). Comparison of three CTAB buffers for plant DNA isolation.
- Bruijns B, Hoekema T, Oomens L, Tiggelaar R, Gardeniers H (2022) Performance of spectrophotometric and fluorometric DNA quantification methods. Analytica 3(3): 371-384.
© 2025 Vidhya Sagar Murugesan. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






