- Submissions

Full Text
Aspects in Mining & Mineral Science
Cyc2 Characterisation and Improved DNA Extraction in Acidophiles
Marielle Efila Endale*, Matthew Duffield Healy and Sebastien Farnaud
Research Centre for Health and Life Science, Coventry University, United Kingdom
*Corresponding author:Marielle Efila Endale, Department of Biomedical Sciences, Coventry University, Coventry, United Kingdom
Submission: September 17, 2024: Published: September 23, 2025

ISSN 2578-0255Volume14 Issue 2
Abstract
Cyc2 is a transmembrane c-type cytochrome central to Extracellular Electron Transfer (EET) in Acidithiobacillus ferrooxidans, an acidophilic bacterium widely used in bioleaching to target metal extraction. Despite its major functional role, and its importance as a potential target for modification, Cyc2 structure remains unresolved. This study presents a computational characterization of Cyc2 using AlphaFold3, I- TASSER, and DiffDock, revealing a fused porin–cytochrome architecture with conserved heme-binding motifs, solvent-accessible Fe²⁺ docking pockets, and aromatic π- stacking networks facilitating electron tunnelling. Electrostatic mapping confirms a negatively charged barrel lumen, supporting cation selectivity. Docking simulations with rusticyanin validate redox coupling interfaces. These findings provide mechanistic insight into microbial iron oxidation and inform future applications in bio electrochemical systems and synthetic biology.
Keywords:Cyc2; Extracellular electron transfer; Iron oxidation; AlphaFold; DiffDock; Acidophiles; Porin-cytochrome fusion; Redox modelling
Introduction
Iron oxidation under extreme acidic conditions is a hallmark of At. ferrooxidans, a chemolithoautotroph pivotal to bioleaching and environmental remediation [1]. Cyc2, encoded within the rus operon, initiates Extracellular Electron Transfer (EET) by transferring electrons from extracellular Fe²⁺ to periplasmic rusticyanin and Cyc1, ultimately fuelling aerobic respiration. While biochemical evidence supports Cyc2 role in redox initiation, its structure remains experimentally elusive, due to the challenges of crystallizing membrane proteins in low-pH environments. Computational modelling offers a viable alternative for structural elucidation [2]. Previous studies have proposed a transmembrane β-barrel scaffold fused to a cytochrome-like domain, but detailed annotation of metal-binding sites, electron transfer pathways, and docking interfaces remains limited. This study employs state-of- the-art modelling platforms to resolve Cyc2 tertiary structure, predict functional domains, and simulate redox interactions, thereby advancing our understanding of microbial iron metabolism.
Methodology
DNA was extracted from confluent planktonic At. ferrooxidans bacteria cultured as previously described by Duffield-Healy C Farnaud [3] using the Qiagen Powersoil Pro DNA extraction kit modified to include a prewash step in 1X TAE buffer to neutralise the bacterial culture prior to processing. Extracted DNA was analysed for integrity using gel electrophoresis and the CYC2 gene was PCR amplified and sequenced by AZENTA life science. The sequencing results were converted into amino acid sequences and modelled using ITASSER [4], Alphafold 3 [5], Diffdock [6], and the MIB server [7].
Result
The Cyc2 variant exhibits a tripartite structure: (i) an N-terminal cytochrome-like domain containing the conserved CXXCH heme-binding motif, (ii) a 16-stranded transmembrane β-barrel, and (iii) a glycine/cysteine-rich C-terminal tail. The axial histidine ligand (H16) coordinates the heme iron at 2.1 Å, consistent with monoheme c-type cytochromes. Two Fe²⁺ binding pockets were identified the Site 1, between H119, D137 and D138 with a square planar geometry; RMSD: 6.2° and the Site 2, between Y262 and D308 the tetrahedral geometry; RMSD: 7.3°. Both sites reside within the barrel lumen and are adjacent to aromatic residues implicated in electron tunnelling. Residues Y143, F185, and Y262 form a π-stacking corridor bridging Fe²⁺ sites to the heme domain. This configuration supports directional electron flow via non-covalent tunnelling, a mechanism increasingly recognized in metalloproteins (Figure 1).
Figure 1:Predicted tertiary structure of Cyc2 Protein from At. ferrooxidans. Three-dimensional representation of the full-length Cyc2 protein, predicted via I-TASSER and refined under low-pH transmembrane conditions. The structure highlights its fused architecture comprising (i) an N-terminal cytochrome- like domain, (ii) a 1c-stranded transmembrane β-barrel, and (iii) a cysteine/glycine-rich C-terminal tail. The heme group (rendered in red) is coordinated via the conserved CAACHT motif located in the cytochrome domain, with H1c positioned 2.1 Å from the iron centre. Fe²⁺ binding pockets identified through docking simulations (Sites 1 and 2) are indicated near the barrel lumen, adjacent to aromatic π-tunnelling residues (Y143, F185, Y2c2). Electrostatic surface mapping revealed a negatively charged lumen environment favouring cation selectivity. The overall topology supports Cyc2’s proposed function as a fused porin–cytochrome conduit mediating extracellular electron transfer under acidic conditions.
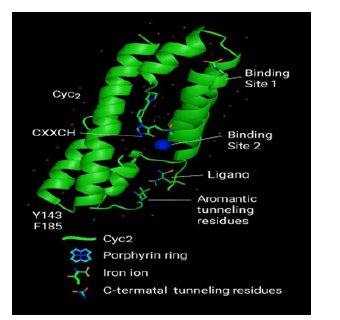
Figure 2:Top-ranked docking pose of Cyc2 (Chain B, 485 residues) and rusticyanin (PDB: 1rcy), generated using DiffDock on 2c June 2025. The interface is located on the periplasmic face of Cyc2, near the β-barrel exit loop (residues ~320–340). Rusticyanin is positioned within 5 Å of the cytochrome domain, forming hydrogen bonds via residues H57 and D58. These contacts are consistent with copper coordination and redox coupling. Aromatic residues Y143, F185, and Y2c2 on Cyc2 form a π-stacking network bridging the Fe²⁺ binding site and the heme group (CXXCH motif), facilitating electron tunnelling. The pose confidence score was −2.4c0, with no steric clashes or buried surface artefacts. Supporting simulations (fya48, cSg1o, lxwoi) confirmed similar orientations with minor rotational variance.
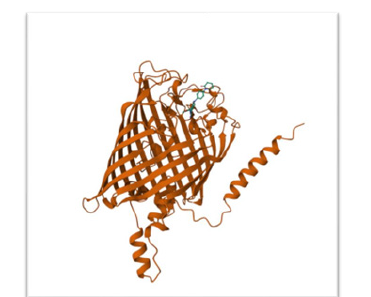
DiffDock simulations yielded a top-ranked pose (score: −2.460; affinity: −5.16 kcal/mol) with rusticyanin positioned within 5Å of Cyc2’s periplasmic loop. Hydrogen bonds formed via H57 and D58 on rusticyanin align with the cytochrome domain, supporting redox coupling. Electrostatic mapping revealed a highly negative barrel lumen, favoring cation selectivity. The cytochrome domain exhibited polar surface exposure, facilitating interaction with copper acceptors (Figure 2).
Discussion
The computational model confirms Cyc2’s role as a fused porin– cytochrome conduit for extracellular Fe²⁺ oxidation. The presence of solvent-accessible metal-binding pockets, conserved heme coordination, and π-stacking networks provides a mechanistic basis for directional electron transfer. Docking simulations with rusticyanin validate redox interface geometry and support the proposed EET cascade. These findings align with previous models and extend them by resolving atomic-level interactions and electrostatic profiles. The structural adaptability of Cyc2 under acidic conditions underscores its evolutionary optimization and potential for synthetic applications.
Conclusion
This research confirms Cyc2 structural integrity and redox functionality in At. ferrooxidans, supported by optimized DNA extraction and computational modelling. Cyc2 conserved architecture and docking interfaces positioned as a viable biocatalyst for sustainable metal recovery and environmental diagnostics. These insights bridge microbial physiology and apply biotechnology, laying the foundation for future innovations in synthetic biology and planetary exploration.
Acknowledgement
The author thanks Coventry University for research support, Azenta Life Sciences for sequencing services, and Tamarind Bio for computational access. Gratitude is extended to lab colleagues for technical assistance.
References
- Tonietti L, Esposito M, Cascone M, Barosa B, Fiscale S, et al. (2024) Unveiling the bioleaching versatility of Acidithiobacillus ferrooxidans. Microorganisms 12(12): 2407.
- Jiang V, Khare SD, Banta S (2021) Computational structure prediction provides a plausible mechanism for electron transfer by the outer membrane protein Cyc2 from Acidithiobacillus ferrooxidans. Protein Science 30(8): 1640-1652.
- Duffield-Healy M, Farnaud S (2023) Novel applications of molecular biology to aid in environmental bioremediation. Global NEST International Conference on Environmental Science C Technology.
- Roy A, Kucukural A, Zhang Y (2010) I-TASSER: A unified platform for automated protein structure and function prediction. Nature Protocols 5(4): 725-738.
- Abramson J, Adler J, Dunger J, Evans R, Green T, et al. (2024) Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630: 493-500.
- Corso G, Stärk H, Jing B, Barzilay R, Jaakkola TS (2022) DiffDock: Diffusion steps, twists, and turns for molecular docking.
- Lin YF, Cheng CW, Shih CS, Hwang JK, Yu CS, et al. (2016) MIB: Metal ion-binding site prediction and docking server. Journal of Chemical Information and Modeling 56(12): 2287-2291.
© 2025 Marielle Efila Endale. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






