- Submissions

Full Text
Annals of Chemical Science Research
Fine-Tuning ChemBERTa-2 for Aqueous Solubility Prediction
Andrew SID Lang1*, Wei Khiong Chong2 and Jan HR Wörner1
1Department of Computing & Mathematics, Oral Roberts University, USA
1Advent Polytech Co., Ltd., Taipao City, Taiwan
*Corresponding author:Andrew SID Lang, Department of Computing & Mathematics, Oral Roberts University, Tulsa, OK, 74171, USA
Submission: May 08, 2023;Published: May 19, 2023

Volume4 Issue1May , 2023
Abstract
Traditional machine-learning techniques for predicting physical-chemical properties often require the calculation and selection of molecular descriptors. Calculating descriptors can be time-consuming and computationally expensive, and there is no guarantee that all relevant and significant features will be captured, especially when trying to predict novel endpoints. In this study, we demonstrate the effectiveness of transformer models in predicting physical-chemical endpoints by fine-tuning the open ChemBERTa-2 model to predict aqueous solubility directly from structure with comparable accuracy to traditional machine-learning techniques, without the need for descriptor calculation and selection. Our findings suggest that transformer models have the potential to provide an efficient and streamlined method for predicting physical-chemical properties directly from molecular structure.
Keywords:Transformer models; ChemBERTa-2; SMILES; Cheminformatics; Physical-chemical property prediction
Introduction
Transformer models are becoming significant tools to support research and development efforts in various areas of chemistry, with many applications based on the open-source BERT [1] and RoBERTa [2] models. These include ChemBERTa [3], MolBERT [4]; SMILES-BERT [5]; and most recently, ChemBERTa-2 [6]. ChemBERTa-2 was trained using masked-language modeling (MLM) and multi-task regression (MTR) on a dataset of 77 million SMILES strings. SMILES, a widely used text representation of molecules, has a straightforward vocabulary with very few grammar rules that encodes each molecule using a sequence of characters that symbolize atoms and bonds [7]. While SMILES has become the dominant language for training chemical transformer models, emerging research suggests that SELFIES, an alternative chemical language, could be a more effective choice for molecular encoding [8].
Recent advancements have shown that foundational models such as the generative pretrained transformer (GPT) can also be adapted to solve various chemistry and materials science tasks by simply prompting them with natural language chemistry-related questions [9,10]. GPT-based models can learn from just a few examples and perform tasks such as classification and regression without modifying their architecture or training methods. This breakthrough demonstrates the potential of large language models (LLMs) to extend beyond their original applications and solve increasingly complex problems in chemistry and other fields.
This paper demonstrates the effectiveness of transformer models, specifically the newly released ChemBERTa-2, in predicting physical-chemical property endpoints with comparable accuracy to standard machine-learning techniques without the need for descriptor calculation and selection. We illustrate this by fine-tuning the ChemBERTa-2 model to predict aqueous solubility, achieving a level of performance like that reported in recent literature [11]. This exemplifies the potential of transformer models to provide a convenient and straightforward method for property prediction directly from structure (SMILES)..
Method
Lowe et al. [11] recently utilized a random forest machine learning technique to predict aqueous solubility on a high-quality curated dataset of experimentally determined solubility values. To compare their results with those of a transformer model, we finetuned the ChemBERTa-2-based model, ChemBERTa-77M-MTR, which is available on Hugging Face [12], using the same dataset. We began by randomly splitting the dataset (N=10207) into training (N=8165), validation (N=1020), and test sets (N=1022) and trained the model using the Trainer class from the transformer’s library with the adamw_torch optimizer. We monitored the performance of the model on the validation set at intervals of ten steps during the training process, enabling us to use the early stopping technique to prevent overfitting while maintaining model accuracy. That is, we ceased training when the validation loss stopped improving.
After optimizing the tuning parameters through our finetuning process, we trained a new transformer model on the original training set from the aqueous solubility paper [11]. We then utilized our newly created transformer model to predict the aqueous solubility of the original test set compounds from the same paper. The code for fine-tuning the ChemBERTa-77M-MTR model and its application to predicting aqueous solubility is available on GitHub [13].
Results
Our results show that by fine-tuning the ChemBERTa-77MMTR model, a chemical language transformer that is based on the Google-developed BERT (Bidirectional Encoder Representations from Transformers) pre-trained natural language processing model, we were able to accurately predict aqueous solubility directly from structure encoded in the language of SMILES. Our performance metrics (R2: 0.822, RMSE: 0.938, MAE: 0.681, ρ: 0.899, p-value: <0.001) are comparable to those of previously published models. Please refer to Table 1 and Figure 1 for a more detailed comparison.
Table 1:Performance Metrics for Random Forest, Transformer Model, and Fine-Tuned Transformer Model.
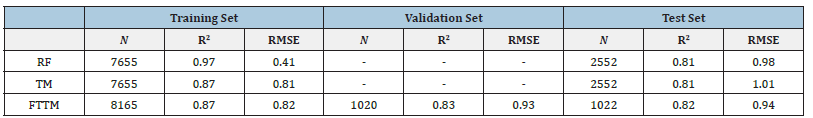
Table Abbreviations: N-Number of points, R2-The coefficient of determination, RMSE-Root mean square error, RFRandom Forest, TM-Transformer model, FTTM-Fine-tuned transformer model.
Figure 1:Test set aqueous solubility – predicted vs. measured for the fine-tuned ChemBERTa-77M-MTR transformer model (ρ: 0.899, p-value: < 0.001) colored by absolute error (AE).
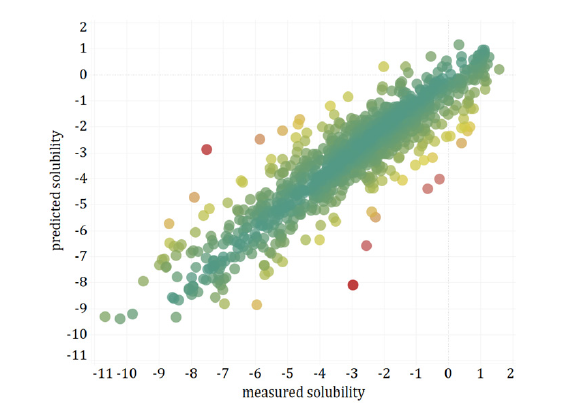
The five molecules with the largest absolute error, see Figure 1,
have the following SMILES (AE):
S=C=NC1C=CC(Br)=CC=1 (3.75),
CN(C)C(=O)NC1=CC(Cl)=C(C=C1)SC(F)(F)Cl (3.77),
NC1C=C(N)C(=CC=1)N=NC1=CC(=CC=C1)N=NC1=CC=C(N)
C=C1N (4.04),
CC1OC(OCC2OC(OC3=CC4OC(=CC(=O)C=4C(O)=C3)C3C=C(O)
C(=CC=3)OC)C(O)C(O)C2O)C(O)C(O)C1O (4.62),
C1CC2=CC3=CC4=CC=CC=C4C=C3C3=CC=CC1=C23 (5.12).
The complexity of these molecules, which feature uncommon functional groups, multiple halogens (chlorine and fluorine), polycyclic structures with multiple nitrogen and oxygen atoms, and intricate polycyclic aromatic hydrocarbon configurations, makes it difficult to predict their solubility values and reflects the model’s lack of knowledge about these compounds.
Discussion
We fine-tuned the ChemBERTa-77M-MTR model to predict aqueous solubility and compared its performance with the results of a previously published random forest model. The results indicate that the fine-tuned transformer model can accurately predict aqueous solubility directly from SMILES, with performance metrics comparable to those found in previously published studies [11]. However, a limitation of the model is that it may have difficulty predicting solubility values for unusual or complex molecules, but this is not necessarily surprising.
This study demonstrates the potential of transformer models in providing an efficient and streamlined method for property prediction directly from structure. This approach does not require descriptor calculation and selection. By not using descriptors, transformer models may seem more difficult to interpret. However, the attention heads of these models learn to assign greater weight to active functional groups in the sequence of tokenized atoms. By leveraging attention mechanisms, it is possible to interpret the model from a physical-chemical perspective. This approach, though beyond the scope of this article, allows one to identify the features of the molecule that contribute most significantly to the predicted property, providing valuable insights into the underlying chemistry, potentially discovering new insights that may be missed when using standard descriptor-based methods [6,14].
Conclusion
The research presented in this paper showcases the promise of transformer models, specifically the recently released ChemBERTa-2, in accurately predicting physical-chemical property endpoints without the need for descriptor calculation and selection. By fine-tuning the ChemBERTa-77M-MTR model to predict aqueous solubility, we achieved comparable performance to previously published models [11]. These results demonstrate the usefulness of transformer models in the field of chemistry. Moreover, this
study highlights the potential of large language models, in general, to tackle complex problems in various fields. Further research is needed to fully explore the capabilities of transformer models in chemistry research and development.Acknowledgments
The authors would like to acknowledge the contributions of Abhik Seal, whose work inspired the authors to carry out this study.
References
- Devlin J, Chang MW, Lee K, Toutanova K (2018) Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:1810.04805.
- Liu Y, Ott M, Goyal N, Du J, Joshi M, et al. (2019) Roberta: A robustly optimized bert pretraining approach. ArXiv preprint arXiv:1907.11692.
- Chithrananda S, Grand G, Ramsundar B (2020) Chemberta: Large-scale self-supervised pretraining for molecular property prediction. ArXiv preprint arXiv:2010.09885.
- Fabian B, Edlich T, Gaspar H, Segler, M, Meyers J, et al. (2020) Molecular representation learning with language models and domain-relevant auxiliary tasks. arXiv preprint arXiv:2011.13230.
- Wang S, Guo Y, Wang Y, Sun H, Huang J (2019) SMILES-BERT: large scale unsupervised pre-training for molecular property prediction. In Proceedings of the 10th ACM international conference on bioinformatics, computational biology and health informatics, pp. 429-436.
- Ahmad W, Simon E, Chithrananda S, Grand G, Ramsundar B (2022) Chemberta-2: Towards chemical foundation models. ArXiv preprint arXiv:2209.01712.
- Weininger D (1988) SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. Journal of chemical information and computer sciences 28(1): 31-36.
- Yüksel A, Ulusoy E, Ünlü A, Deniz G, Doğan T (2023) SELFormer: Molecular representation learning via SELFIES language models. ArXiv preprint arXiv:2304.04662.
- Jablonka KM, Schwaller P, Ortega-Guerrero A, Smit B (2023) Is GPT-3 all you need for low-data discovery in chemistry? ChemRxiv. Cambridge: Cambridge Open Engage.
- Ramos MC, Michtavy SS, Porosoff MD, White AD (2023) Bayesian Optimization of catalysts with in-context learning. arXiv preprint arXiv:2304.05341.
- Lowe CN, Charest N, Ramsland C, Chang DT, Martin TM, et al. (2023) Transparency in modeling through careful application of OECD’s QSAR/QSPR principles via a curated water solubility data set. Chemical Research in Toxicology, 36(3): 465-478.
- DeepChem (DeepChem) (2022) Hugging Face.
- Lang ASID, Chong WK, Worner JHR (2023) LCW-Fine-Tuning-ChemBERTa-2 (Version 1.0.0).
- Payne J, Srouji M, Yap DA, Kosaraju V (2020) BERT learns (and teaches) chemistry. ArXiv preprint arXiv:2007.16012.
© 2023 Andrew SID Lang. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






