- Submissions

Full Text
Annals of Chemical Science Research
Applications of Targeted Protein Degradation in Drug Discovery
Xue H1#, Chen Y1# and Jin J1,2,3*
1Life Sciences Institute, Zhejiang University, Hangzhou, Zhejiang, China
2The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, China
3Cancer Center, Zhejiang University, Hangzhou, Zhejiang, China
#These authors contributed to this article equally
*Corresponding author: Jin J, Life Sciences Institute, Zhejiang University, Hangzhou, Zhejiang, China Email: jianping_jin@zju.edu.cn
Submission: April 04, 2021;Published: May 06, 2021

Volume2 Issue4May, 2021
Abstract
Proteostasis is a process that controls protein abundance in living cells in order to maintain healthy proteome at both cell and organ levels. In eukaryotes, the ubiquitin-proteasome pathway and the lysosomal network are two main systems for this purpose. Since the first attempt was tried to design drugs using protein degradation principles in 2001, rapid progress has been made over the past 20 years. In this review article, we will summary a few approaches to design protein degradation drugs, including PROTACs, Homo-PROTACs, Opto-PROTACs, PHOTACs, CLIPTACs, ATTEC, AUTACs, as well as LYTACs, and briefly discuss their current progression.
Introduction
Human genome contains about 22,000 genes, of which around 4,500 are considered to be human disease-linked. Most conventional drugs, including antibodies and small molecules, target proteins to cure human diseases. Druggable proteins often contain active centers where traditional small molecule drugs can efficiently access. Unfortunately, ~85% of the human proteome is regarded to be undruggable, that is to say, these proteins lack active pockets and are beyond the reach of conventional drugs. However, recent developments in drug discovery field have paved ways to design drugs targeting these undruggable proteins. Human cells have evolved two systems to promote turnover of unwanted or damaged proteins, as well as injured organelles. One of such systems is via the proteasome and the other is through the lysosome. In this short review, we will summary the recent progress in drug design approaches based on protein degradation principles.
Drug Design Approaches Based on the Ubiquitin-Proteasome System (UPS)
The ubiquitin-proteasome system degrades unwanted proteins to maintain normal cellular activities in eukaryotes. Protein ubiquitylation (also termed ubiquitination), the first part of UPS, consists of three steps of enzymatic reactions catalyzed by ubiquitinactivating enzyme E1, ubiquitin conjugating enzyme E2, and ubiquitin ligase E3 [1]. One of the consequences of protein ubiquitylation is protein turnover via the 26S proteasome [1]. Protein ubiquitylation is a specific process which is largely determined by over 600 ubiquitin ligases. Many components of UPS, including ubiquitin ligases, ubiquitin-specific proteases, and subunits of the proteasome, are drug targets. For example, several proteasome inhibitors, like bortezomib, carfilzomib, and ixazomib, have been approved by the U.S. Food and Drug Administration (FDA) as drugs to treat multiple myeloma. Moreover, small molecule ligands of ubiquitin ligases have started to be employed in developing drugs based on protein degradation.
Proteolysis-Targeting Chimeras (PROTACs)
PROTACs are small molecule-based heterobifunctional compounds with three elements: a ligand binding to the protein of interest (POI), a second ligand specifically interacting with a ubiquitin ligase, and a linker connecting both ligands [2-4]; (Figure 1). Upon binding to both POI and the ubiquitin ligase, PROTACs can drag POI close to the ubiquitin ligase to trigger POI ubiquitylation and then degradation via the 26S proteasome [5]. The first PROTAC was created under the collaboration between Raymond Deshaies’ laboratory and Craig Crews’ group [2]. They built a chimeric molecule, named Protac-1, which contains the IκB-α phosphodegron specifically recognized by the SCFβ-TrCP ubiquitin ligase and ovalicin that covalently binds to methionine aminopeptidase-2 (MetAP-2). With Protac-1, SCFβ-TrCP was able to recruit MetAP-2 and to drive its ubiquitylation and degradation [2]. The first generation PROTACs were peptide-based including Protac-2 and -3, targeting estrogen receptor (ER) and androgen receptor (AR) respectively [6]. However, due to the lack of cell permeability, these PROTACs are hard to be delivered into living cells. This disadvantage limited their applications greatly. To overcome this issue, the first cellpermeable PROTAC named Protac-4 was developed [7]. Protac-4 contains an artificial ligand AP21998 that binds to the mutant F36V FKBP12 proteins and an ALAPYIP peptide that recruits the CRL2VHL ubiquitin ligase [7,8]. In addition, a poly-D-arginine sequence derived from the HIV TAT protein was coupled to the peptidic E3 recruiting moiety to confer cell permeability and to resist nonspecific proteolysis. The resulting PROTAC can target and degrade the mutant FKBP12 proteins at 25 μM in cells [7]. From then on, cell-penetrating peptide has been frequently used in peptide-based PROTACs [9,10].
Figure 1:Traditional PROTACs consist of an E3 ligand, a ligand of protein substrate and a linker connecting both parts. PROTACs tie substrate to the E3 ligase, leading to ubiquitylation and degradation of protein substrate.
However, several limitations imply that the peptide-based PROTACs might not be good drug candidates, such as high molecular weight, potential immunoreaction, relatively poor cell permeability and metabolic instability [4,11]. With the requirement of better drug-like PROTACs, the first entire small-molecule PROTAC based on nutlin, a ligand of MDM2 which is a ubiquitin ligase promoting p53 ubiquitylation and turnover [12], and a non-steroidal AR ligand, connected by a polyethylene glycol-based linker was developed [13]. This novel compound can trigger the degradation of AR by selective recognition. Compared with peptidic PROTACs, small molecule-based PROTACs own many distinct advantages, such as stability, biodistribution and potency. Indeed, small molecule ligands of several E3 ligases (CRBN, VHL, MDM2, clAP1 etc) fueled the progress of PROTACs. Perhaps, one of the breakthroughs is the discovery of thalidomide as a ligand of Cereblon, one of the substrate receptors in the Cullin4-RING ubiquitin ligases [14]. Thalidomide and its derivatives, including lenalidomide and pomalidomide etc, function as molecular glues to promote proteolysis of Ikaros family proteins, CK1α, GSPT1, SALL4 and p63 etc. Since then, thalidomide and its derivatives, especially lenalidomide, have been widely used in constructing PROTACs [15-18]. At the similar time, another ligand of substrate receptor in CRL2VHL ubiquitin ligase was discovered under the cooperation of Crews’ laboratory and Yuichi Hashimoto’s group [19,20]. Based on the ligand of Celeblon and VHL, a series of PROTACs targeting BRD4 and FKBP12 were synthesized [21- 23], igniting the passion for drug discovery on PROTACs. Indeed, the first two clinical trials with PROTACs aiming at prostate and breast cancers produced favorable results. In recent years, several new PROTAC-related technologies such as Homo-PROTACs [24- 26], opto-PROTACs [27,28], photochemically targeting chimeras (PHOTACs) [29,30], as well as click-formed proteolysis targeting chimeras (CLIPTACs) [31], have also been created.
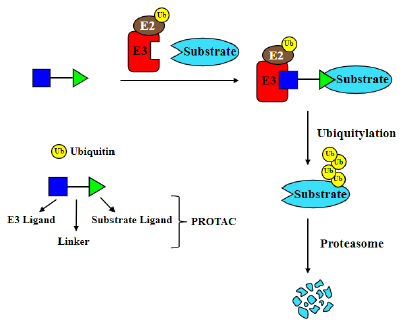
Homo-PROTACs
Homo-PROTACs is a method to apply PROTACs to induce dimerization of a given ubiquitin ligase, and then to trigger its autoubiquitylation and suicide-type proteolysis in living cells [24-26]; (Figure 2). For instance, two ligands of VHL, VH032 and VH298, were connected by a linker to create homo-PROTACs which were able to degrade VHL efficiently [24]. The most active compound, CM11, could dimerize VHL and induce neddylationand proteasome-dependent self-degradation of VHL in different cell lines. More interestingly, CM11 could degrade VHL in a highly isoform-dependent manner. Using a similar strategy, homo- PROTACs were constructed by linking two pomalidomide molecules to target CRBN [26]. One of such compounds, 15a, was able to degrade CRBN by autoubiquitylation and proteasome-dependent turnover. In addition, it could also inhibit pomalidomide-dependent proteolysis of Ikaros family proteins and antagonized the antiproliferative effects of IMiDs on multiple myeloma cells.
Figure 2:Homo-PROTACs use both warheads to hijack E3 ubiquitin ligases, resulting in the degradation of ubiquitin ligases.
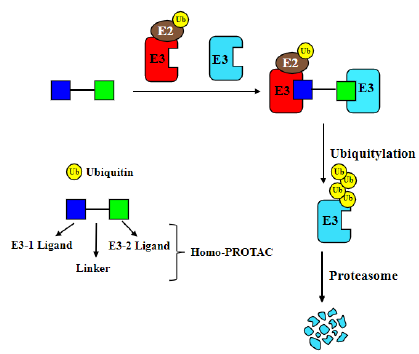
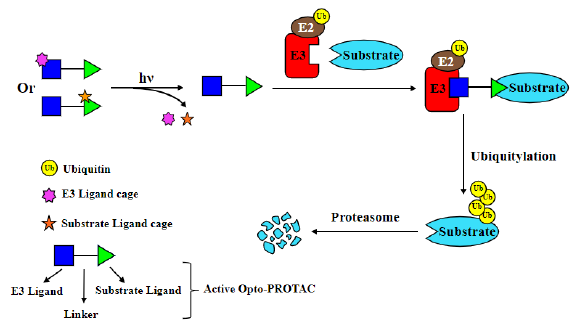
Opto-PROTACs
Opto-PROTACs are light-activatable PROTACs [26,27]; (Figure 3). First, caged PROTACs were created by placing photolabile protecting groups on either substrate ligand or E3-recruiting ligand parts of PROTACs. Then light was applied to activate PROTACs to trigger protein turnover [27]. Using this strategy, opto-dBET1 and opto-dALK were synthesized by installing a photolabile caging group on pomalidomide to degrade BET1 and ALK under a light-inducible manner, respectively. Initially, the pomalidomide molecule on these Opto-PROTACs remains inert. They could be activated and gain degradation ability after illuminated with light of specific wavelength (365 nm), due to releasing the photolabile caging group [28].
Figure 3:Opto-PROTACs have the photolabile group to confine their capabilities, and can be activated by light.
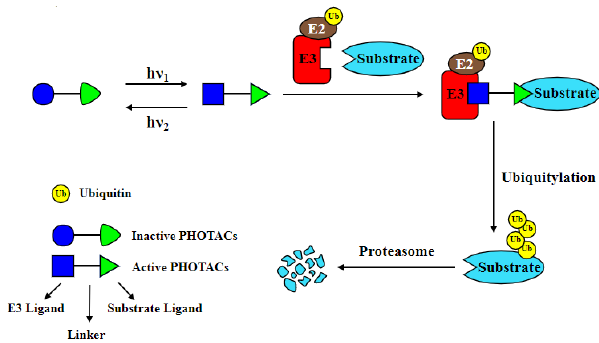
PHOTACs
PHOTACs belong to the other version of light-inducible PROTACs (Figure 4). However, unlike opto-PROTACs which are photocaged PROTACs, PHOTACs are photo-switchable PROTACs. In principle, a photoswitch is added in PHOTACs, allowing them to be controlled by the light in a spatio-temporal manner [29,30]. Azobenzenes are chosen to be the photoswitch for their easily tunable photothermal actuations. By coupling azobenzenes to any of the three components of PROTACs, PHOTACs exhibit little activity and less toxic in dark or under long wavelength light but are activated by the blue-violet light (380-440 nm) or pulse irradiation to induce a conformation change in the azo-linkage, leading to the degradation of targeted proteins. PHOTACs have been applied to degrade either FKBP12 or BET family proteins, including BRD2, BRD3, and BRD4.
Figure 4: PHOTACs are combined with a photoswitch which makes them activated or silenced by light.
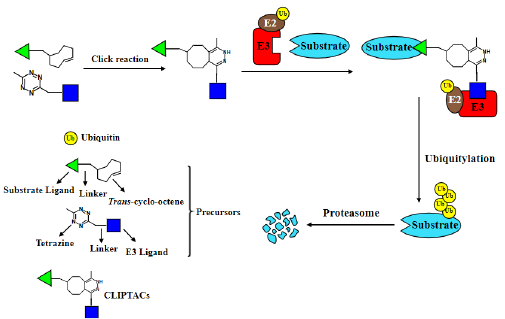
CLIPTACs
CLIPTACs are made from two smaller precursor molecules which can be connected into active PROTACs by a bioorthogonal click reaction in living cells [31]; (Figure 5). In this application, Lebrau et al. (2016) employed the tetrazine-tagged thalidomide as an E3-recruiting ligand and trans-cyclooctene-tagged warheads were used to recognize POIs. Two CLIPTACs were successfully created to target oncoproteins BRD4 and ERK1/2 for ubiquitin- and proteasome-dependent degradation intracellularly. Precursors are small, thus infiltrate cells more easily. When reaching inside cells, they form CLIPTACs through a click reaction, resulting in degradation of targeted proteins[31]. PROTACs and related technologies provide alternative tools for targeting undruggable disease proteins which lack feasible pockets where traditional small molecules can bind, such as scaffold proteins, signaling adaptors, and transcriptional factors. PROTACs have shown vigorous capability of protein turnover both in cells and in animals. Arvinas, the first PROTAC technology-based drug company, has demonstrated clinical efficacy of two PROTACs, ARV-110 for prostate cancer patients and ARV-471 for treatments of breast cancer in Phase 1 clinical trials. However, the traditional PROTACs also exhibit a few disadvantages. They often have large molecular mass, therefore, might be hard to enter cells. Moreover, undesirable ligase-mediated off-target effects pose a significant risk and are not yet well understood. Furthermore, systemic administration of PROTACs might target POIs in both tumor and normal cells, resulting in unexpected adverse effects, and might potentially lead to uncontrolled degradation of the POIs in any cells, e.g., normal cells versus cancer cells. In this case, alternative PROTAC-related approaches could solve some of the issues. However, these applications still need more studies to prove their efficacy in drug discovery.
Figure 5: CLIPTACs are formed by two precursors through the click reaction in cells, and activated afterwards.
Drug Design Methods Based on Lysosome-Dependent Protein Degradation
As the PROTAC-related technologies develop, more protein degradation-associated methods have also emerged. The UPS can’t handle all kinds of proteolytic events, like degradation of damaged organelles, cell membranes, extracellular proteins as well as large protein aggregates. Thus, drug design approaches based on lysosome-dependent degradation pathways have come into sight especially in recent several years. Several protein turnover pathways have intrinsic connections with the lysosome. One of such pathways is endocytosis which could degrade membrane proteins and extracellular materials, including nutrients, proteins, solute molecules as well as extracellular pathogens; Another pathway depends on autophagy which is a self-degradative process to eliminate aggregated or misfolded proteins, to remove damaged organelles, such as mitochondria, endoplasmic reticulum or peroxisome, and to eradicate intracellular pathogens in eukaryotic cells. Autophagy is triggered by a family of ubiquitin-like proteins, such as ATG12, LC3 proteins as well as GABARAP members [32,33], and has less selectivity of targeted proteins. Up to now, several methods have been created to trigger degradation events via the lysosome.
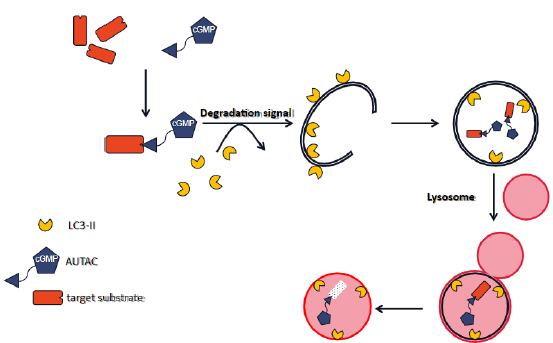
Autophagy-Targeting Chimeras (AUTACs)
AUTACs are cargo-specific degraders based on selective autophagy to target a wide range of substrates including cytosolic proteins as well as organelles for degradation via the autophagylysosome system (Figure 6). Takahashi et al. [34] generated this method based on a principle that endogenous S-guanylation (cGMP modification) can be found on invasive bacteria which was demonstrated to enhance substrates’ ubiquitylation and subsequent degradation through autophagy. Basically, a specific binder of the interested targets is attached by a linker to a guanine tag where S-guanylation forms. As a proof-of-principle, they fused a HaloTag (HT) to EGFP to make a EGFP-HT fusion protein which can be specifically recognized by a cGMP-containing HaloTag ligand (cGMP-HTL). Then by evaluating the cGMP-modified EGFP-HT level, dot-like EGFP structures and co-localization of EGFP dots with LC3B in the system, they confirmed that S-guanylation leads to EGFP degradation through Atg5-mediated autophagy. However, this primary version of AUTACs also showed limitations like low reaction efficiency between the cGMP-modified ligand and the HTtagged protein. Besides, undesired side effects induced by cGMPactivated downstream regulators could also appear. To solve these problems, Takahashi et al. [34] modified the guanine tag by removing negative charge and cGMP activity-related structures to develop a S-guanylation mimicking tag called p-fluorobenzylguanine (FBnG), which was later shown to be superior to the original version with similar EGFP phenotypes but accelerated reaction between ligands and HaloTag. Further studies using specific binders as warhead towards targeted proteins also proved the feasibility of FBnG as a degradation signal in drug discovery, although there are still some limitations like the nuclear-localized targeted substrates are shown with lower degradation efficiency compared to the cytoplasmic ones.
Figure 6:AUTACs are designed based on selective autophagy. They are cargo-specific degraders targeting cytosolic proteins as well as organelles for degradation via the autophagy-lysosome system.
Another type of substrates that can be targeted by AUTACs is damaged organelle, especially mitochondria. Mitophagy is a well-known process of damaged mitochondria clearance through autophagy. Therefore, Takahashi et al. [34] wondered whether this S-guanylation modification system could also induce mitophagy. A fusion protein between EGFP-HT and the mitochondrial outer membrane protein 25 was expressed stably in HeLa cells. Then with the treatment of a FBnG ligand (FBnG-HTL), a co-localization of mitochondria and LC3B but no alterations to mitochondrial protein levels were observed, suggesting that S-guanylation alone was not enough in this case. However, mitochondrial protein levels reduced markedly when knocking down OPA1 to inhibit mitochondrial inner membrane fusion and to release mitochondrial fragments, indicating that FBnG-HTL enhanced the clearance efficiency of fragmented mitochondria. More interestingly, CCCP-induced destroyed mitochondrial membrane potential was partially restored by FBnG-HTL, along with attenuated release of caspase activator cytochrome c from damaged mitochondria, which was canceled by autophagy inhibitor bafilomycin A1 as well as knocking out of Atg5, suggesting a cytoprotective role of this autophagybased system for mitochondria.
Dysfunctional mitochondria accumulation leads to a genetic disease called DS with few therapeutic approaches. Considering the potential role of AUTACs in mitochondrial clearance and biogenesis, Takahashi et al (2019) applied it to DS patient-derived fibroblast cells. Consistently, in this cell context with more fragmented mitochondrial, AUTAC4, one of the AUTACs, was able to trigger mitophagy to remove damaged mitochondria and restore the mitomembrane potential, further shedding light on the therapeutic role of AUTACs [34].
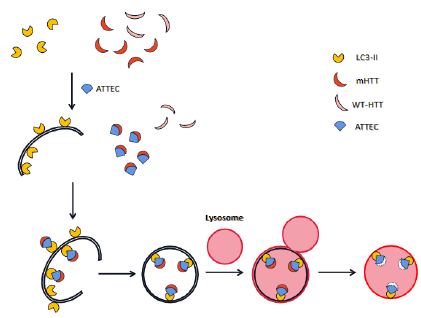
Autophagosome-Tethering Compound (ATTEC)
ATTEC is constructed to link targeted proteins to LC3, a ubiquitin-like protein involved in autophagy initiation [35,36]; (Figure 7). Therefore, targeted proteins could be attached to phagophores for eventual turnover via the autophagy-lysosome machinery. This approach was applied to degrade large protein aggregates and could be a potential therapeutic treatment for neurodegenerative diseases, such as Huntington disease (HD). To search for small molecules suitable for ATTEC, Li et al. (2019) developed a small molecule-based microarray screen and identified four compounds that function as molecular glues between LC3 and mutant huntingtin (mHTT), a causative gene of HD, but not wild-type huntingtin (wtHTT). Moreover, they found that these compounds induced the formation of phagophore engulfing mHTT inside and subsequent lysosome fusion, autolysosome formation and final mHTT degradation. Furthermore, using various models including patient-derived cells and animal models like HD Drosophila models or HD knock-in mouse models, they showed that these compounds specifically targeted mHTT, but not wtHTT, for autophagic degradation and rescued HD-related phenotypes observed in these models. Finally, they confirmed that mHTT was degraded through Atg5-mediated autophagy [35].
Figure 7:ATTEC is created to connect targeted proteins directly to LC3 for degradation via the autophagylysosome system.
In comparison with AUTACs, ATTEC seems to provide much clearer mechanism, since ATTEC employs LC3 ligands to induce protein degradation via the autophagy-lysosome system. Given the fact that ATG12 and GABARAP proteins, another group of ubiquitinlike proteins are also involved in the initiation of autophagosome formation like LC3 proteins, it is conceivable that similar strategy could be employed to develop ATG12- or GABARAP-binding ligands to design ATTECs to drive proteins degradation through autophagy.
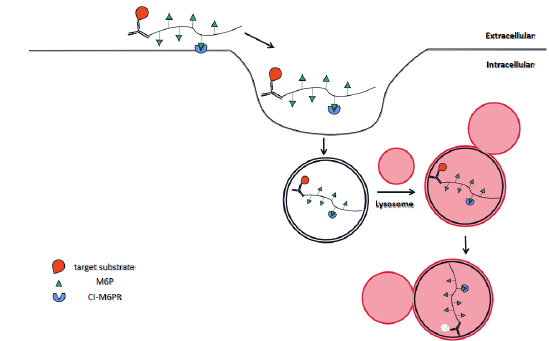
Lysosome-Targeting Chimeras (LYTACs)
Apart from large aggregates, extracellular proteins are also beyond the reach of the ubiquitin-proteasome system, which makes the lysosome system more attractive in targeted proteins degradation. LYTACs were designed specifically to target extracellular proteins like secreted proteins and membraneassociated ones by utilizing the interaction between cell-surface lysosome-targeting receptors (LTRs) and their ligands [37] (Figure 8). LTRs are well-known for the role of trafficking extracellular proteins to the lysosomes. In this case, Banik et al. (2020) focused on a LTR called cation-independent mannose-6-phosphate receptor (CI-M6PR, also known as IGF2R), a transmembrane glycoprotein, which is able to recruit proteins harboring N-glycans capped with mannose-6-phosphate (M6P) residues and then transport them to lysosomes for degradation via endocytosis. In this approach, they designed a complex consisting of a targeted protein specific antibody attached to a synthetic polyM6Pn which serves as the ligand for CI-M6PR, achieving the goal of recognizing extracellular targeted proteins via specific antibody and delivering them to lysosomes for degradation through the interaction between M6P and its receptor on the cell surface at the same time [37]. Potentially, this approach could be used to degrade extracellular and membrane proteins through the lysosome. More excitingly, this LYTACs system was proved to degrade membrane-associated cancer-related proteins like EGFR, CD71 and PD-L1, together with the ability to antagonize the escape of endogenous recycle proteins from the lysosome degradation system.
Figure 8:LYTACs were invented specifically to promote degradation of extracellular and membrane proteins via the endocytosis-lysosome pathway.
Discussion
Nowadays, protein degradation-based approaches start to show more and more promising outcomes in drug inventions. However, many questions still need to be answered. One big question is drug resistance. Small molecular inhibitors often gain resistance, especially in cancer patients, mainly due to mutations of POIs. Whereas protein degradation drugs could gain drug resistance against different components [38,39]. For example, studies have shown that PROTACs can be disarmed resulting from mutations and then inactivation of ubiquitin ligases. Similarly, drugs constructed based on other PROTAC-like methods and lysosomebased degradation approaches could also obtain resistance due to the inactivation in components along these two proteostasis pathways. Further studies are needed to fully survey the nature of these new drug design approaches.
Acknowledgement
This work was partly supported by grants from National Natural Science Foundation of China (NSFC) (31970734), the Fundamental Research Funds for the Central Universities (2019QN81006), and startup funds from the Life Science Institute, Zhejiang University, China.
References
- Swatek KN, Komander D (2016) Ubiquitin modifications. Cell Res 26(4): 399-422.
- Sakamoto KM, Kim KB, Kumagai A, Mercurio F, Deshaies RJ, et al. (2001) Protacs: Chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc Natl Acad Sci USA 98(15): 8554-8559.
- Chen YL, Jin JP (2020) The application of ubiquitin ligases in the PROTAC drug design. Acta Biochimica Biophysica Sinica 52(7): 776-790.
- Toure M, Crews CM (2016) Small-molecule protacs: New approaches to protein degradation. Angew Chem Int Edit 55(6): 1966-1973.
- Corson TW, Aberle N, Crews CM (2008) Design and applications of bifunctional small molecules: Why two heads are better than one. Acs Chem Biol 3(11): 677-692.
- Sakamoto KM, Kim KB, Verma R, Ransick A, Stein B, et al. (2003) Development of protacs to target cancer-promoting proteins for ubiquitination and degradation. Mol Cell Proteomics 2(12): 1350-1358.
- Schneekloth JS, Fonseca FN, Koldobskiy M, Mandal A, Deshaies R, et al. (2004) Chemical genetic control of protein levels: Selective in vivo targeted degradation. J Am Chem Soc 126(12): 3748-3754.
- Yang W, Rozamus LW, Narula S, Rollins CT, R Yuan, et al. (2000) Investigating protein-ligand interactions with a mutant FKBP possessing a designed specificity pocket. J Med Chem 43(6): 1135-1142.
- Montrose K, Krissansen GW (2014) Design of a PROTAC that antagonizes and destroys the cancer-forming X-protein of the hepatitis B virus. Biochem Bioph Res Co 453(4): 735-740.
- Chu TT, Gao N, Li QQ, Chen PG, Yang FX, et al. (2016) Specific knockdown of endogenous tau protein by peptide-directed ubiquitin-proteasome degradation. Cell Chem Biol 23(4): 453-461.
- Zou YT, Ma DH, Wang YY (2019) The PROTAC technology in drug development. Cell Biochem Funct 37(1):21-30.
- Brooks CL, Gu W (2003) Ubiquitination, phosphorylation and acetylation: The molecular basis for p53 regulation. Curr Opin Cell Biol 15(2): 164-171.
- Schneekloth AR, Pucheault M, Tae HS, Crews CM (2008) Targeted intracellular protein degradation induced by a small molecule: En route to chemical proteomics. Bioorg Med Chem Lett 18(22): 5904-5908.
- Ito T, Ando H, Suzuki T, Ogura T, Hotta K, et al. (2010) Identification of a primary target of thalidomide teratogenicity. Science 327(5971): 1345-1350.
- Lu G, Middleton RE, Sun HH, Naniong M, Christopher J Ott, et al. (2014) The myeloma drug lenalidomide promotes the cereblon-dependent destruction of ikaros proteins. Science 343(6168): 305-309.
- Gandhi AK, Kang J, Havens CG, Conklin T, Ning Y, et al. (2014) Immunomodulatory agents lenalidomide and pomalidomide co-stimulate T cells by inducing degradation of T cell repressors Ikaros and Aiolos via modulation of the E3 ubiquitin ligase complex CRL4(CRBN). Brit J Haematol 164(6): 811-821.
- Kronke J, Fink EC, Hollenbach PW, MacBeth KJ, Slater N Hurst, et al. (2015) Lenalidomide induces ubiquitination and degradation of CK1 alpha in del(5q) MDS. Nature 523(7559): 183-188.
- Donovan KA, An J, Nowak RP, Yuan JTC, Emma C Fink, et al. (2018) Thalidomide promotes degradation of SALL4, a transcription factor implicated in Duane Radial Ray syndrome. Elife, p. 7.
- Buckley DL, Van Molle I, Gareiss PC, Tae HS, Michel J, et al. (2012) Targeting the von hippel-lindau E3 ubiquitin ligase using small molecules to disrupt the VHL/HIF-1 alpha Interaction. J Am Chem Soc 134(10): 4465-4468.
- Buckley DL, Gustafson JL, Van Molle I, Roth AG, Tae HS, et al. (2012) Small-molecule inhibitors of the interaction between the E3 ligase VHL and HIF1 alpha. Angew Chem Int Edit 51(46): 11463-11467.
- Winter GE, Buckley DL, Paulk J, Roberts JM, Souza A, et al. (2015) Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348(6241): 1376-1381.
- Lu J, Qian YM, Altieri M, Dong HQ, Wang J, et al. (2015) Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem Biol 22(6): 755-763.
- Zengerle M, Chan KH, Ciulli A (2015) Selective small molecule induced degradation of the BET bromodomain protein BRD4. Acs Chem Biol 10(8): 1770-1777.
- Maniaci C, Hughes SJ, Testa A, Chen WZ, Lamont DJ, et al. (2017) Homo-PROTACs: Bivalent small-molecule dimerizers of the VHL E3 ubiquitin ligase to induce self-degradation. Nat Commun, p. 8.
- Girardini M, Maniaci C, Hughes SJ, Testa A, Ciulli A (2019) Cereblon versus VHL: Hijacking E3 ligases against each other using PROTACs. Bioorgan Med Chem 27(12): 2466-2479.
- Steinebach C, Lindner S, Udeshi ND, Mani DC, Kehm H, et al. (2018) Homo-PROTACs for the chemical knockdown of cereblon. Acs Chem Biol 13(9): 2771-2782.
- Naro Y, Darrah K, Deiters A (2020) Optical control of small molecule-induced protein degradation. J Am Chem Soc 142(5): 2193-2197.
- Liu J, Chen H, Ma LN, He ZX, Wang D, et al. (2020) Light-induced control of protein destruction by opto-PROTAC. Sci Adv 6(8).
- Wang ZW, Liu Y, Zhu XQ (2020) PhotoPROTACs: A novel biotechnology for cancer treatment. Trends Cell Biol 30(10): 749-751.
- Reynders M, Matsuura BS, Berouti M, Simoneschi D, Marzio A, et al. (2020) PHOTACs enable optical control of protein degradation. Sci Adv 6(8).
- Lebraud H, Wright DJ, Johnson CN, Heightman TD (2016) Protein degradation by in-cell self-assembly of proteolysis targeting chimeras. Acs Central Sci 2(12): 927-934.
- Levine B, Kroemer G (2019) Biological functions of autophagy genes: A disease perspective. Cell 176(1-2): 11-42.
- Parzych KR, Klionsky DJ (2014) An overview of autophagy: Morphology, mechanism, and regulation. Antioxid Redox Signal 20(3): 460-473.
- Takahashi D, Moriyama J, Nakamura T, Miki E, Takahashi E, et al. (2019) AUTACs: Cargo-specific degraders using selective autophagy. Mol Cell 76(5): 797-810.
- Li Z, Wang C, Wang Z, Zhu C, Li J, et al. (2019) Allele-selective lowering of mutant HTT protein by HTT-LC3 linker compounds. Nature 575(7781): 203-209.
- Li Z, Zhu C, Ding Y, Fei Y, Lu B (2020) ATTEC: A potential new approach to target proteinopathies. Autophagy 16(1): 185-187.
- Banik SM, Pedram K, Wisnovsky S, Ahn G, Riley NM, et al. (2020) Lysosome-targeting chimaeras for degradation of extracellular proteins. Nature 584(7820): 291-297.
- Zhang L, Gillis BR, Vijay P, Shen Y (2019) Acquired resistance to BET-PROTACs (Proteolysis-Targeting Chimeras) caused by genomic alterations in core components of E3 ligase complexes. Mol Cancer Ther 18(7): 1302-1311.
- Ottis P, Palladino C, Thienger P, Britschgi A, Heichinger C, et al. (2019) Cellular resistance mechanisms to targeted protein degradation converge toward impairment of the engaged ubiquitin transfer pathway. ACS Chem Biol 14(10): 2215-2223.
© 2021 Jin J. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






