- Submissions

Full Text
Research in Medical & Engineering Sciences
Development of High-Content Adult Sensory Neuron Gene Silencing Screening Assay to Study Mechanisms of Neurite Growth
Phillip Canete1, David Do1, Richard Lie1, Grace Woodruff2 and Gunnar Poplawski1,3*
1Department of Neurosciences, University of California, USA
2Department of Cellular and Molecular Medicine, University of California, USA
3Center for Immunotherapy and Precision Immuno-Oncology (CITI), Lerner Research Institute at the Cleveland Clinic, USA
*Corresponding author:Gunnar HD Poplawski, Assistant Professor, Poplawski Laboratory, Center for Immunotherapy and Precision Immuno-Oncology (CITI), Lerner Research Institute, Cleveland Clinic, USA
Submission: October 05, 2023;Published: October 16, 2023

ISSN: 2576-8816Volume10 Issue4
Abstract
Genome wide RNA-sequencing has become an important tool to identify potential candidate genes involved in axon regeneration following spinal cord or peripheral nerve injury. One of the major benefits but also challenges of such big data approaches is the identification of hundreds to thousands of potential candidate genes that ultimately have to be subjected to further screening methods, before investigation in in vivo models of axon regeneration is feasible. Thus, we have developed a protocol to efficiently silence mRNA expression in adult DRG neurons in the 384-well plate format, which allows for rapid neurite-morphology screening of candidate genes. We compared two widely used lipid-based transfection reagents Lipofectamine2000 and RNAiMAX and investigated their efficacy on silencing of EGFP-expression in adult sensory neurons from Fischer 344 rats that ubiquitously express EGFP. We assessed mRNA knockdown by EGFP-expression in the neuronal cell body and determined overall cell health via assessment of neurite outgrowth. Here we present that both transfection reagents produce comparable results leading to EGFP silencing efficiencies of up to 60% in adult sensory neurons with minimal impact on neuronal survival. We validate our assay by showing increased neurite growth via PTEN mRNA knock-down. This assay allows for rapid and cost-efficient screening of hundreds to thousands of candidate genes and to evaluate their impact on neuronal morphology and axon regeneration.
Keywords:Lipid-based transfection; Dorsal root ganglion (DRG) neuron; siRNA; Axon regeneration; 384-well plate; Gene silencing screen
Introduction
Transcriptomic and proteomic screens have become an integral tool to study mechanism of axon regeneration following nerve injury [1-4]. These powerful, big-data approaches offer a system wide understanding of mechanisms involved in axon regeneration. Performing multi-omic screens on a specific model of axon regeneration can significantly deepen the understanding of the mechanisms involved. Often times these screens are performed in multiple conditions, thus leading to thousands of significantly differentially expressed genes. One of the major challenges of these multi-omic approaches is the wealth of data one has to analyze to ultimately identify the key players that are mediating the regenerative state [5].
In vitro screening of candidate genes investigating axon regeneration offers a pathway to successfully narrow down extensive gene lists to a manageable count for further investigation in in vitro or in vivo models of spinal cord regeneration [6,7]. It is important to select the appropriate type of screening assay in order to aid in proper candidate selection. Considerations should be included.
A. The neuronal cell type,
B. The method of gene manipulation
C. The method of transfection,
D. The multi-well plate format
E. The method of readout,
F. The equipment needed, and
G. The available funds to plan and carry out successful in
vitro screens.
Neuronal cell type: When utilizing mammalian primary neuronal cultures, one is mostly restricted to embryonic and postnatal ages. One exception are sensory neurons isolated from dorsal root ganglia (DRGs), which can be cultured as early as embryonic day 10 (E10) [8,9], as well as in an adult state, without the need of neurotropic support [10]. DRG neurons can be cultured for an extended amount of time in vitro while retaining many of their immunocytochemical and physiological characteristics [10- 14]. They further provide an excellent model to study the cellintrinsic mechanisms of axonal regeneration, since the expression of regeneration associated genes can be induced prior to tissue harvesting by a peripheral nerve injury [6,15-17]. DRG neurons thus been instrumental in identifying and validating candidate genes involved in mechanism of axon regeneration that have emerged from multi-omic screens [1-4].
Method of gene manipulation: Genetic manipulations resulting in gain or loss of function of candidate genes can in general be achieved by candidate gene over-expression or genesilencing. Both overexpression plasmid libraries as well as siRNA/shRNA libraries are commercially available. For economic reasons, we decided to utilize siRNA molecules for a gene silencing approach, since small scale libraries are much more affordable than overexpression, or shRNA libraries.
Method of transfection: The transfection options fall into two basic categories: viral and non-viral. While viral transduction is more effective, it is also more time-consuming, expensive and requires additional safety precautions [18,19]. Non-viral transfection techniques include direct DNA delivery via injection or biolistics [20], sonoporation [21], electroporation, chemical vectors, cationic polymers and lipofection [22-24]. While electroporation has become very efficient in the recent years, it requires expensive equipment and consumables. Lipofection transfers DNA or RNA molecules into the cell in a lipid carrier that fuses with the cell membrane [25]. The advantage of lipofection is its cost-efficiency and ease of application. DNA/RNA molecules can be pre-incubated with lipofection reagent and lipid-nucleic acid complexes can be added to the already adherent neurons. Lipofection leads to high yield transfection in dividing cells but poses difficulties in transfection postmitotic neurons [26]. In this study we are comparing two widely used lipofection reagents Lipofectamine2000 (for DNA and siRNAs) and RNAiMAX (specifically for siRNA-molecules).
Multi-well plate format: The source of the optical cell culture plate can be an important consideration when performing highcontent screening assays investigating the effect of candidate gene silencing on neurite morphology. The physical characteristics of multi-well tissue-culture plates, such as type of plastics and materials used, as well as charge-altering surface treatments (e.g., plasma or corona), can have a significant impact on neuronal attachment, survival and on other neuromorphological parameters [27,28]. Those aspects of a multi-well plate seem to become more prevalent, with increasing number of wells, resulting in increased surface to well ratio, in which surface tensions and electrostatic properties become more predominant and can hence directly impact neuronal properties [29]. Considering all these aspects we decided to utilize 384-well plates, in which surface tensions are reasonable and which offer the ability to simultaneously screen libraries of several thousand siRNA molecules but can still be easily managed with handheld multichannel/stepper pipettes.
Method of readout: DRG neurons were derived from
transgenic adult Fischer 344 rats that ubiquitously express EGFP.
We are utilizing the EGFP-fluorescent signal as a direct readout
for successful neuron transfection, mRNA-silencing and protein
knock-down. Utilizing EGFP expression as readout has two major
advantages.
a) EGFP expression can easily be detected by a fluorescent
microscope and quantified by image analysis software and
more importantly correlates directly with the amount of EGFPprotein;
b) Reduction of EGFP has no known impact on neuronal
function. For determining axon regeneration, neuronal health
and other neuromorphological parameters that could be
affected by candidate gene silencing, we are utilizing the
neuronal marker βIII-tubulin immunofluorescent staining
combined with automated neurite tracing algorithms as
readout [30-32].
Equipment needed: We utilized multi-channel/multi-stepper pipettes, 384-well optical plates, an automated imaging system (ImageXpress, Molecular Devices) and software for automated image analysis and quantification of neuro-morphological parameters (MetaXpress, Molecular devices). An important additional feature of the image analysis software is the ability to measure EGFP fluorescence signal intensity specifically within the neuronal cell bodies, that co-labeled with βIII-tubulin (Custom module editor, Molecular Devices). To run this assay one can use any fluorescent microscope with an automated stage followed by automated image analysis. Software packages that are specifically designed for neuro-morphological screens in 384-well plate format are available as freeware [30].
Available funds: We designed this assay to be as economical viable as possible, so small labs with limited funds can perform meaningful gene silencing screens. With this optimized protocol, a screen for 500 candidate genes can be repeated several times in triplicates for under 10k USD.
The following data presents the development and validation of a functional assay that was designed under all these considerations. We developed a step-by-step protocol that is easy to follow, allowing scientists with no high-content screening background to gather meaningful data within a few weeks. The described assay can easily be scaled to screen genome wide siRNA libraries.
Result
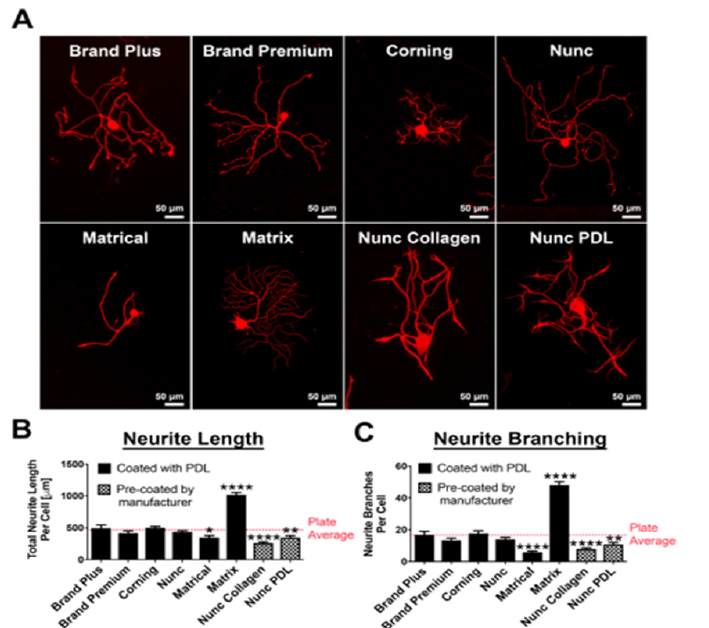
Differences in 384-well plate manufacturing directly influences neurite morphology of adult DRG neurons
To establish a baseline for neurite morphological parameters, such as neurite outgrowth, initiation and branching as well as neuronal survival, we compared 384-well plates from different manufacturers (Figure 1). Surprisingly we found significant differences in neurite length and branching depending on the manufacturer of the 384-well optical tissue culture plate. Culturing DRG neurons in plates from Matrix resulted in a 100% increase (p<0.0001, un-paired t-test; Figure 1B), while plates from Matrical showed a ~30% reduction in neurite outgrowth compared to average neurite outgrowth over all plates (p<0.05, un-paired t-test; Figure 1B). Even more prominent effects of the plate manufacturer were observed on neurite branching. Matrix plates increase branching by almost 2-fold, while Matrical plates showed a 65% reduction in branches compared to plate averages (p<0.0001, un-paired t-test) (Figure1C). Similar plate-dependent neurite outgrowth properties were observed with hIPSC derived neurons (Figure S1), indicating that differences in plate manufacturing have a general influence on neurite morphology from different neuronal sources. We subsequently decided to move forward with 384- well plates from Nunc, which displayed average value of neurite outgrowth and branching (Figure 1B, C) and showed even cell distribution (data not shown).
Figure 1:Neurite outgrowth and branching of adult dorsal root ganglion neurons is influenced by plate type
manufacturer in 384-well plates.
(A) βIII-tubulin staining of representative DRG neurons in 384-well plates from different manufacturers. All plates
coated with either Poly-D-Lysine or pre-coated by the manufacturer (Nunc collagen and Nunc PDL). Scale bars:
50μm.
(B, C) Quantification of (B) total neurite length per cell (μm) and (C) Number of neurite branches per cell in 384-
well plates from different manufacturers compared to plate average. n=3 independent experiments with n=4 wells/
experiment. Mean+SEM. p<0.0001 one-way ANOVA with Bonferroni’s posthoc, *p<0.05, **p<0.01, ****p<0.0001.
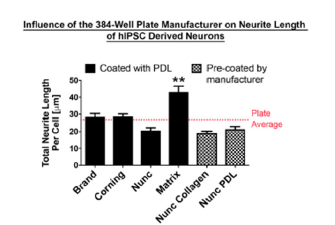
Supplementary Figure 1:Neurite Outgrowth of human induced pluripotent stem cells is influenced by plate type manufacturer on 384-well plate Comparison of neurite outgrowth per cell (μm). All plates were coated with either Poly-D-Lysine or pre-coated by the manufacture. n=3 independent experiments with n=10 wells/experiment. Mean+SEM. p<0.0001 one-way ANOVA with Bonferroni’s posthoc, **p<0.01.
Stimulatory and inhibitory substrates can be utilized to alter neuronal morphology and thereby increase the dynamic detection range
When performing high-content screens, it is critical to work within a high dynamic range to ensure that positive and negative hits fall within the detection spectrum. When the readout is a neurite morphological analysis, such as neurite outgrowth, decreasing the minimum and increasing the maximum growth limits can expand the dynamic range. To establish these limits in our assay, we show a dose dependent increase in neurite length with the addition of laminin (Figure 2A,B) and dose dependent reduction of neurite length with the addition of chondroitin sulfate proteoglycans (CSPGs, Figure 2C,D). It is critical that these reagents are used in concentration ranges, in which neurite morphological parameters are not reaching a plateau state (>30μg/ml laminin/ CSPG; Figure 2A,B), which could potentially dominate the neuromorphological outcome and hence overpower the effect of the candidate gene manipulation. It is advised to identify EC50 and IC50 values, which represent reagent concentrations, where 50% of the maximum stimulatory (EC50) or inhibitory (IC50) effect is observed (Figure 2). Laminin stimulation promoted neurite growth by a 10- fold maximum (EC50=5μg/ml; 5-fold increase in neurite growth; Figure 2A middle panel, B); whereas CSPG inhibition reduced neurite growth up to a maximum of 28% (IC50 = 15.0μg/ml; 14% decrease in neurite growth; Figure 2C middle panel, D). To establish an assay that aims to screen for positive manipulators of neurite outgrowth (e.g., axon regeneration), it can be advantageous to utilize conditions with reduced neurite outgrowth, such as coating with inhibitory substrates (e.g., CSPGs) at their IC50.
Figure 2:Neurite outgrowth and branching of adult DRG neurons is stimulated upon laminin treatment and
inhibited upon CSPG treatment in 384-well plates.
Neuronal morphology is assessed via the neuronal marker βIII-tubulin. (A) DRG neurons without Laminin (top
panel), and upon Laminin stimulation (lower panels). 50% (middle panel) and 100% (bottom panel) neurite
outgrowth stimulation is shown.
(B) Dose response curve of total neurite length per cell (μm) in response to increasing concentrations of Laminin.
50% of neurite outgrowth stimulation (EC50) is achieved by 5μg/ml laminin.
(C) DRG neurons without CSPGs (top panel), and upon CSPG inhibition (lower panels). 50% (middle panel) and
100% (bottom panel) neurite outgrowth inhibition is shown.
(D) Dose response curve of total neurite length per cell (μm) in response to increasing concentrations of CSPGs. 50%
of neurite outgrowth inhibition (IC50) is achieved by 15μg/ml CSPGs. n=3 independent experiments with n=5 wells/
experiment. Mean±SEM. Scale bars: A, 100μm; C, 50μm.

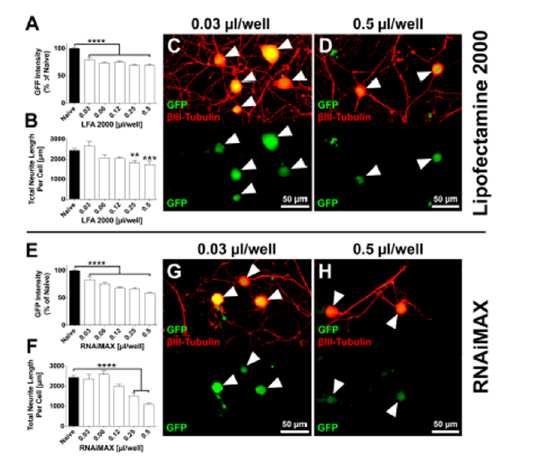
Titration of transfection reagent: increasing amounts of transfection reagent lead to reduced EGFP-expression, but also increased toxicity
To assess transfection efficiency as well as gene silencing efficacy, we utilized transgenic Fischer433 rats that ubiquitously express EGFP. This model allows for EGFP-mRNA silencing and subsequent EGFP (protein) reduction, which (1) can be used as direct readout of gene silencing efficiency via fluorescent intensity of EGFP specifically in neuronal cell bodies in culture and (2) has no impact on neuronal function. We tested a range of 0.03-0.5μl lipid-based transfection regent per well, while keeping the siRNA concentration constant (2.5pmol/well) and determined EGFPexpression and neurite length after 48 hours in vitro (Figure 3), whereas neurite length functions as a determinant of neuronal health. EGFP expression was significantly reduced compared to control conditions for all concentrations of transfection reagents applied for both Lipofectamine2000 and RNAiMAX (p<0.0001, One-way ANOVA with Dunnett’s post-hoc; Figure 3A,E). We further observed a trend of reduced EGFP expression with the application of increased amounts of transfection reagent, but we also saw a significant reduction in neurite growth for concentrations above 0.12μl/well for both Lipofectamine2000 and RNAiMAX (p<0.0001, One-way ANOVA with Dunnett’s post-hoc; Figure 3B,F). We thus decided to use 0.12μl of transfection reagent per well for both Lipofectamine2000 and RNAiMAX for further optimizations to achieve maximum EGFP reduction with minimal impact on neurite growth.
Figure 3:EGFP expression and neurite outgrowth are reduced in response to increasing concentrations of
lipofection reagent.
Transfection of DRG neurons with either with (A-D) Lipofectamin 2000 (LFA2000), or (E-H) RNAiMAX. Both (A)
LFA2000 and (E) RNAiMAX show significant reduction of EGFP intensity with the application of 0.03-0.5μl/well.
p<0.0001, one-way ANOVA with Dunnett’s posthoc, ****p<0.0001. (B, F) Neurite length is significantly reduced
at lipofection reagents concentrations above 0.12μl/well. p<0.0001, one-way ANOVA with Dunnett’s posthoc,
**p<0.01, ***p<0.001, ****p<0.0001. n=3 independent experiments with n=5 wells/experiment. Mean+SEM. (C, D)
Example images showing DRG neurons transfected with EGFP-siRNA at 0.03μl/well LFA2000 (lowest concentration)
and 0.5μl/well LFA2000 (highest concertation) respectively. (G, H) Example images showing adult DRG neurons
transfected with GFP-siRNA at 0.03μl/well RNAiMAX (lowest concentration) and 0.5μl/well RNAiMAX (highest
concertation) respectively. Scale bars: 50μm. Arrowheads indicate position of neuronal bodies.
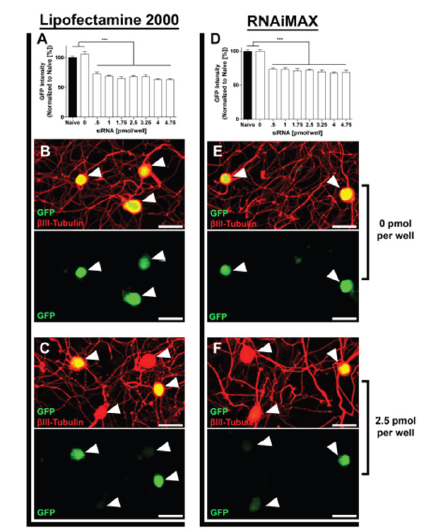
Titration of siRNA molecules: varying amounts of siRNA molecules have no significant impact on EGFP expression and neuronal morphology
With a constant concentration of transfection reagent (0.12μl/ well), we investigated EGFP expression with varying siRNA concentrations ranging from 0.5 to 4.75pmol EGFP siRNA per well (corresponds to 8 to 80nM; Figure 4). All tested concentrations showed similar levels (~25% average/cell) of EGFP knockdown in neuronal cell bodies for both Lipofectamine2000 (Figure 4A) and RNAiMax (Figure 4D) (p<0.0001, One-way ANOVA with Dunnett’s post-hoc). We did not observe any change in neurite growth related to the amount of siRNA molecules applied (data not shown). We thus concluded that a concentration within the middle of our tested range (between 2-3pmol/well; 30-50nM) would be optimal for screening of unverified siRNA molecules, to account for siRNA molecules with less silencing efficacy and potential off-target effects.
Figure 4:EGFP Expression is unaffected by siRNA concentration. (A) EGFP Intensity normalized to naive [%] after transfection with EGFP siRNA [pmol/well] and 0.12μl/well of Lipofectamine 2000. (D) EGFP Intensity normalized to naive [%] after transfection with EGFP siRNA [pmol/well] and 0.12μl/well of RNAiMAX. (B) EGFP Expression after treatment with 0pmol/well of siRNA and 0.12μl/well of Lipofectamine 2000. (E) EGFP Expression after treatment with 0pmol/well of siRNA and 0.12μl/well of RNAiMAX. (C) EGFP Expression after treatment with 2.5pmol/well of siRNA and 0.12μl/well of Lipofectamine 2000. (F) EGFP Expression after treatment with 2.5pmol/well of siRNA and 0.12μl/well of RNAiMAX. Arrowheads indicate position of neuronal bodies. n=3 independent experiments with n=10 wells/experiment. Mean+SEM. p<0.0001, one-way ANOVA with Dunnett’s posthoc, ***p<0.001. Scale bar: 50μm.
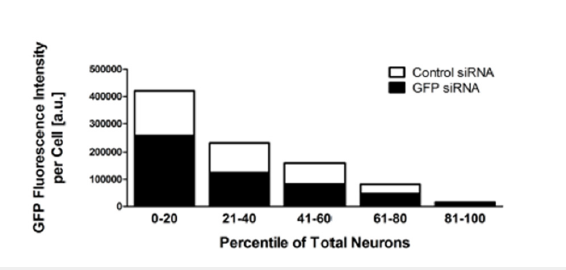
Optimized screening conditions reduce EGFP expression by half in 45% of neurons
In earlier optimization steps we observed an average reduction of EGFP expression by 25% including successfully transfected neurons as well as un-transfected neurons. To more accurately determine the siRNA transfection efficiency as well as EGFP silencing efficiency, we investigated EGFP fluorescence on a single cell basis. We found that our optimized transfection conditions of 2.5pmol siRNA and 12.5μl Lipofectamine2000 transfection reagent per well (40nM) reduced EGFP fluorescence by 50% in at least 45% of all DRG neurons (Figure 5). The EGFP-siRNA transfected neurons with the highest EGFP knockdown displayed up to 60% reduction in EGFP fluorescent signal intensity.
Figure 5:EGFP fluorescence intensity is reduced by at least 50% in 45% of all neurons. Cells were sorted by individual EGFP fluorescence intensity and binned into 5 groups based on their fluorescent signal. Groups from left to right represent each twentyest percentiles of all cells from highest to lowest EGFP signal intensity. White bars represent fluorescent intensity of control siRNA transfected neurons, and black bars represent the corresponding fluorescent signal intensity of GFP siRNA transfected neurons.
PTEN mRNA silencing significantly increases axon growth
Figure 6:PTEN siRNA transfection significantly increases axon growth in adult DRG neurons. (A-C) Example images of adult DRG neurons transfected with PTEN siRNA, Allstars siRNA and control siRNA with optimized conditions, respectively. (D) Neurite outgrowth normalized to control siRNA. n=3 independent experiments with n=10 wells/experiment. Mean+SEM. p<0.0001, one-way ANOVA with Dunnett’s posthoc, ***p<0.001. Scale bar: 50μm.
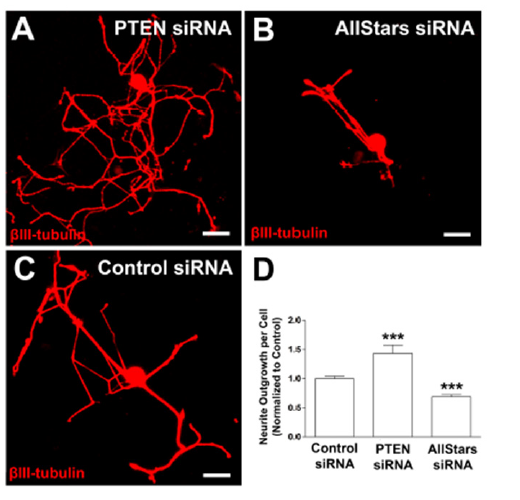
To determine if our assay is able to detect a significant change in neurite length via silencing of a single gene, we applied the optimized transfection conditions to siRNA molecules that have previously been validated to alter neurite length. Reduced Phosphatase and tensin homolog (PTEN) expression has been shown to increase axon extension in vivo [33,34] and in vitro [35]. We consequently utilized siRNAs targeting PTEN (Figure 6A) to investigate if neurite length will significantly increase upon knock-down of PTEN in our assay. We further targeted cell growth and survival mechanisms (AllStars positive control siRNA, Qiagen, Figure 6B) to determine if we can observe a significant reduction in axon growth. As a reference we utilized a non-targeting control siRNA (Life Technologies, Figure 6C). Transfection with siRNA molecule targeting PTEN significantly increased neurite outgrowth by 40% (p<0.001, un-paired t-test; Figure 6D), while transfection with the AllStars siRNA reduced in neurite outgrowth by 30% compared to a non-targeting control siRNA (p<0.001, un-paired t-test; Figure 6D). These results indicate the level of siRNA transfection and subsequent gene silencing is sufficient to significantly alter neuro-morphological parameters, which can be used as a successful readout for screening purposes.
Experimental Procedures
Animal care
All procedures involving animals were carried out in strict adherence to guidelines provided by The Guide for the Care and Use of Laboratory Animals (The Institute of Laboratory Animal Resources, 2011), The Public Health Service Policy on Humane Care and Use of Laboratory Animals (NIH, 1986), The Animal Welfare Act/Regulations and subsequent amendments (PL 89-544), and The Veterans Health Administration Handbook 1200.07 “Use of Animals in Research” (2011); VA San Diego Healthcare System (VASDHS) Research Services Policy 01 section 151-04 (Institutional Animal Care and Use Committee, IACUC) and VASDHS IACUC Policy 03 (Pre and Post-procedural Care of Laboratory Rodents). The animal use protocol was approved by the VASDHS IACUC (Protocol number 14- 008). F344-Tg (UBC-EGFP) F455Rrrc rats were obtained from the Rat Resource & Research Center and maintained at the VA hospital in San Diego, CA. All rats were maintained on a 12-hr light/dark cycle and given ad libitum access to food and water.
Plate type analysis
384 well plates ((Brand Plus and Brand Premium (BrandTech Scientific, Inc. , Essex, CT), Corning (Corning, Corning, NY), Matrical (Spokane, Washington), Matrix, Nunc, Nunc Collagen, Nunc PDL (Thermo Scientific, Waltham, MA)) were precoated with Poly- D-lysine hydrobromide (PDL, 20μg/ml in water, Sigma-Aldrich, St. Louis, MO) overnight at room temperature, washed 3 times with sterile water and air dried then coated with either laminin (0-200μg/ml (Sigma-Aldrich, St. Louis, MO)) or Chicken CSPG (0- 200μg/ml (Bellirica, MA)) in DPBS (Life Technologies, Carlsbad, CA) for 4 hours at room temperature, and washed 3 times with media (DMEM/F12 with GlutaMAX (Life Technologies, Carlsbad, CA),10% FBS (Gemini, Sacramento, CA) , 1x B27 (Invitrogen, Grand Island, NY)).
DRG culture
DRGs were removed, stripped of their roots and collected in HBSS (Life Technologies, Carlsbad, CA) on ice. DRGs were washed once with HBSS, digested with HBSS+0.25% collagenase XI (SigmaAldrich, St. Louis, MO)+5mg/ml Dispase (Worthington, Lakewood, NJ) for 30 min at 37 °C, washed once with media, triturated in media, and plated onto a 384 well tissue culture treated plates at 100-400 DRG neurons in 70μl media per well. After plating, the cells were allowed to settle for 30 minutes followed by incubation at 37 °C and 5% CO2 until ready for transfection.
Immunohistochemistry
Neurons were fixed with 4% Formaldehyde (Fisher Scientific, Pittsburg, PA) in DPBS for 20 min at room temperature, washed 3 times with PBS, blocked and permeabilized in DPBS + 0.25% Triton X-100 (Fisher Scientific, Pittsburg, PA) + 5% horse serum for one hour at room temperature. Cells were then incubated with primary antibody mouse anti-ßIII-tubulin (1:1000, Promega, Fitchburg, WI) in DPBS +.25 Triton X-100 + 5% horse serum for 1.5 hours at room temperature, washed 3 times with DPBS + Triton X-100 + 5% horse serum, incubated with secondary antibody Alexa 647 anti-mouse (1:1000 Life Technologies, Carlsbad, CA) and 4′,6-diamidino-2- phenylindole dihydrochloride (DAPI; [1μg/mL]; Sigma-Aldrich, St. Louis, MO) in DPBS + Triton X-100 + 5% horse serum for one hour at room temperature, washed 3 times with DPBS and imaged with ImageXpress (Molecular Devices, Sunnyvale, CA). Data was analysed with MetaXpress (Molecular Devices), graphs and statistical analysis were done with Prism (GraphPad, La Jolla, CA).
Transfection
The transfection reagent (Lipofectamine 2000 and RNAiMAX, Invitrogen, Carlsbad, CA) was diluted in Optimem (Life Technologies, Carlsbad, CA) at 0-100μl/ml. siRNA molecules EGFP-siRNA and control-siRNA, (Life Technologies, Carlsbad, CA) Block-iT Alexa Flour Red Fluorescent Control (Thermo Scientific, Waltham, MA), Allstars Death siRNA (Qiagen), and Rn_PTEN_7 Flexitube siRNA (Qiagen) were diluted in optimem at 0-0.95uM. The mixtures were incubated for 20 min at room temperature. 5μl of reagent mix was combined with 5μl of siRNA mix and incubated for 20 min at room temperature. 10μl of the combined mix was added per well (384-well plate). The plate was preincubated at room temperature for 30 min and transferred to conditions of 37 °C in 5% CO2. 3 hours after transfection, the media was exchanged with (DMEM/F12 w/Glutamax,10% FBS, 1x B27, penicillin and streptomycin (Life Technologies, Carlsbad, CA; 1000U per 10mg/ ml final concentration). All DRG neurons were then cultured at 37 °C in 5% CO2 for 48 hours.
Human iPSC generation and culture
Human iPSC derived neurons were derived and purified as previously described (47). In brief, after iPSC generation from patient derived fibroblasts (47), iPSCs were differentiated to NPCs and further to neurons as previously described (48). 3×105 FACS-purified TRA1-81+ cells were seeded onto 3×10cm plates that were seeded the previous day with 5×105 PA6 cells. On day 11, cells were dissociated with Accutase and ~5 × 105 CD184+CD15+CD44−CD271- NPCs were FACS-purified and plated onto poly-ornithine/laminin-coated plates and cultured with bFGF. NPCs were differentiated by removal of bFGF from the media. After 3 weeks of differentiation, cells were dissociated with Accutase and CD24+CD184-CD44- cells were purified. FACS was performed with a FACSAria II (BD Biosciences). Purified neurons were then plated in 384-well plates (Matrix, Thermofischer) at 2500 cells per well in 50μl media (DMEM: F12 + Glutamax, 0.5x N2, 0.5x B27 (both from Life Technologies), 1x P/S (Millipore), 0.5mM dbCAMP (Sigma- Aldrich, St. Louis, MO), 20ng/μl BDNF and 20ng/μl GDNF (both from Peprotech)). Plates were pre-coated overnight at 37 °C with poly-ornithine (40μg/ml in water, Sigma-Aldrich, St. Louis, MO), washed 3 times with sterile water, subsequently coated with either rat or monkey myelin (10μg/ml in PBS) overnight at 4 °C, washed 3x with PBS, coated with laminin (5μg/ml in media) for 4 hours at room temperature, washed 3x with media before plating of cells. Cells were cultured at 37 °C in 5% CO2 for 48 hours.
Data and statistical analysis
Neurite outgrowth, EGFP and Block-iT fluorescent intensity were analyzed with MetaXpress (Molecular Devices, Sunnyvale, CA). Results are represented as mean ± standard error of the mean (SEM) with at least 5 wells per condition. Prism (GraphPad, La Jolla, CA) was used to calculate the SEM and statistical significance of difference between groups, evaluated by one-tailed Student’s t-test or one way ANOVA with Bonferroni’s or Dunnett’s Test to determine significance of a condition *p<0.05; **p<0.01; ***p<0.001 against a control.
Discussion
The powerful approach of unbiased high throughput siRNA screening has been described recently [36], allowing for rapid investigation of candidate gene function in biological pathways, such as axon regeneration. Here we developed a high-content assay to screen siRNA libraries for the identification of modulators of axon regeneration in adult DRG neurons [1-4].
One of the challenges in designing a gene silencing screen in postmitotic cells, such as neurons, is achieving suitable transfection rates, without having to rely on labor and cost intensive transfection methods such as electroporation or viral based transfection [37]. We thus investigated several widely used lipid-based transfection reagents and determined their efficiency in silencing EGFP expression in adult DRG neurons derived from rats that ubiquitously express EGFP. The initial panel included Lipofectamine 3000 (Invitrogen) [38], MessengerMax, RNAiMAX (Invitrogen) [39,40]. and Lipofectamine2000 (Invitrogen) [26], as well as self-delivering siRNAs (Dharmacon) that do not require additional transfection reagents [41]. Initial investigation of all reagents showed the most reliable EGFP protein knock-down for Lipofectamine2000 and RNAiMAX, on which we subsequently focused our investigations. Utilizing Lipofactamine2000 and RNAiMAX we achieved similar EGFP knock-down compared to self-delivering siRNA molecules (Accell from Dharmacon), which can be added directly to the growth media [41] (unpublished observations), but for a fraction of the cost. Thus, Accell is a viable option for screening a limited number of siRNAs in DRG neurons but becomes economically nonviable for larger screens.
RNAiMAX, which was specifically developed for the transfection of siRNA molecules showed higher transfection efficiency and less toxicity in certain cell types [42,43]. We show that in adult DRG neurons, Lipofectamine 2000 and RNAiMAX result in comparable gene silencing efficiencies with no significant difference in toxicity. We thus recommend the usage of Lipofectamine 2000 over RNAiMax if co-transfection of a reporter plasmid is desired [39,40].
We decided to use EGFP fluorescence as a proxy for determining the efficiency of gene silencing, since it allows for a direct readout of successful protein reduction without relying on immunolabeling [44,45]. Boudes and colleagues reported an average reduction of EGFP expression up to 40% in DRG neurons using single cell electroporation [44]. This method of transfection, however, is timely and cost inefficient for high content screening. Our assay using inexpensive reagents is able to reduce the average EGFP expression up to 25% in DRG neurons. This magnitude of EGFP reduction closely replicates the decrease in EGFP signal observed in embryonic hippocampal neurons also transfected with lipofectamine 2000 and EGFP siRNA [45]. In addition, our method demonstrates at least a 50% reduction of EGFP expression in up to 45% of DRG neurons, an improvement over previous optimization of lipofection that reported efficiencies between 20-30% in other primary neurons [26].
To establish sensitivity of our assay to detect significant morphological changes of a single target gene, we targeted PTEN to promote neurite growth [35]. We observed 40% increase in neurite growth compared to non-targeting control siRNA, significantly more than a previously reported 25% increase with similar methods [35]. We could further significantly reduce neurite growth by 30% with the application of Allstar control siRNA (Qiagen) that targets cell maintenance pathways. We thus successfully designed an assay that has the potential to identify genes that either promote or reduce neurite growth of adult DRG neurons in 384-well format in vivo. A detailed step-by-step protocol can be found in the supplemental materials of this manuscript.
Conclusion
We developed an assay that allows for rapid screening of commercial siRNAs libraries to evaluate the impact of candidate gene silencing on neuro-morphological parameters, such as axon regeneration in adult DRG neurons. These screening conditions can be applied to siRNA libraries of unknown efficacy and the impact of specific siRNA molecules on neurite-morphology can be easily assessed by βIII-tubulin staining and automated neurite tracing [30]. Positive or negative regulators of neurite-morphology have to be consequently validated in follow up experiments in detail. This protocol allows laboratories with access to a standard fluorescent microscope with automated stage to effortlessly screen hundreds of candidate genes within a short time frame and thus make informed decisions on target selection, before conducting further in vivo or in vivo studies.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgement
Not applicable
Permission to Reuse and Copyright
Not applicable
References
- Chandran V, Coppola G, Nawabi H, Omura T, Versano R, et al. (2016) A systems-level analysis of the peripheral nerve intrinsic axonal growth program. Neuron 89(5): 956-970.
- Costigan M, Befort K, Karchewski L, Griffin RS, D'Urso D, et al. (2002) Replicate high-density rat genome oligonucleotide microarrays reveal hundreds of regulated genes in the dorsal root ganglion after peripheral nerve injury. BMC Neurosci 3: 16.
- Michaelevski I, Ruder YS, Rozenbaum M, Medzihradszky KF, Shalem O, et al. (2010) Signaling to transcription networks in the neuronal retrograde injury response. Sci Signal 3(130): ra53.
- Stam FJ, MacGillavry HD, Armstrong NJ, de Gunst MCM, Zhang Y, et al. (2007) Identification of candidate transcriptional modulators involved in successful regeneration after nerve injury. Eur J Neurosci 25(12): 3629-3637.
- Poplawski GHD, Kawaguchi R, Niekerk EV, Lu P, Mehta N, et al. (2020) Injured adult neurons regress to an embryonic transcriptional growth state. Nature 581(7806): 77-82.
- Poplawski G, Ishikawa T, Brifault C, Kubli CL, Regestam R, et al. (2018) Schwann cells regulate sensory neuron gene expression before and after peripheral nerve injury. Glia 66(8): 1577-1590.
- Poplawski GHD, Lie R, Hunt M, Kumamaru H, Kawaguchi R, et al. (2018) Adult rat myelin enhances axonal outgrowth from neural stem cells. Sci Transl Med 10(442): eaal2563.
- Dulbecco R (1970) Topoinhibition and serum requirement of transformed and untransformed cells. Nature 227(5260): 802-806.
- Rhoades RW, Enfiejian HL, Chiaia NL, GJ Macdonald, Miller MW, et al. (1991) Birthdates of trigeminal ganglion cells contributing axons to the infraorbital nerve and specific vibrissal follicles in the rat. J Comp Neurol 307(1): 163-175.
- Malin SA, Davis BM, Molliver DC (2007) Production of dissociated sensory neuron cultures and considerations for their use in studying neuronal function and plasticity. Nat Protoc 2(1): 152-160.
- Baccaglini PI, Hogan PG (1983) Some rat sensory neurons in culture express characteristics of differentiated pain sensory cells. Proc Natl Acad Sci U S A 80(2): 594-598.
- Gold MS, Dastmalchi S, Levine JD (1996) Co-expression of nociceptor properties in dorsal root ganglion neurons from the adult rat in vivo. Neuroscience 71(1): 265-275.
- Reid G, Flonta ML (2001) Physiology. Cold current in thermoreceptive neurons. Nature 413(6855): 480.
- Wood JN, Winter J, James IF, Rang HP, Yeats J, et al. (1988) Capsaicin-induced ion fluxes in dorsal root ganglion cells in culture. J Neurosci 8(9): 3208-3220.
- Neumann S, Skinner K, Basbaum AI (2005) Sustaining intrinsic growth capacity of adult neurons promotes spinal cord regeneration. Proc Natl Acad Sci U S A 102(46): 16848-16852.
- Neumann S, Woolf CJ (1999) Regeneration of dorsal column fibers into and beyond the lesion site following adult spinal cord injury. Neuron 23(1): 83-91.
- Richardson PM, Issa VM (1984) Peripheral injury enhances central regeneration of primary sensory neurones. Nature 309(5971): 791-793.
- Berger G, Durand S, Fargier G, Nguyen XN, Cordeil S, et al. (2011) APOBEC3A is a specific inhibitor of the early phases of HIV-1 infection in myeloid cells. PLoS Pathog 7(9): e1002221.
- Kato T, Date T, Murayama A, Morikawa K, Akazawa D, et al. (2006) Cell culture and infection system for hepatitis C virus. Nat Protoc 1(5): 2334-2339.
- Dib Hajj SD, Choi JS, Macala LJ, Tyrrell L, Black JA, et al. (2009) Transfection of rat or mouse neurons by biolistics or electroporation. Nat Protoc 4(8): 1118-1126.
- Lin CR, Chen KH, Yang CH, Cheng JT, Sheen Chen SM, et al. (2010) Sonoporation-mediated gene transfer into adult rat dorsal root ganglion cells. J Biomed Sci 17(1): 44.
- Al Dosari MS, Gao X (2009) Nonviral gene delivery: principle, limitations, and recent progress. AAPS J 11(4): 671-681.
- Inoue T, Krumlauf R (2001) An impulse to the brain--using in vivo Nat Neurosci 4 Suppl: 1156-1158.
- Washbourne P, McAllister AK (2002) Techniques for gene transfer into neurons. Curr Opin Neurobiol 12(5): 566-573.
- Tseng YC, Mozumdar S, Huang L (2009) Lipid-based systemic delivery of siRNA. Adv Drug Deliv Rev 61(9): 721-731.
- Dalby B, Cates S, Harris A, Ohki EC, Tilkins ML, et al. (2004) Advanced transfection with Lipofectamine 2000 reagent: primary neurons, siRNA, and high-throughput applications. Methods 33(2): 95-103.
- Ranella A, Barberoglou M, Bakogianni S, Fotakis C, Stratakis E (2010) Tuning cell adhesion by controlling the roughness and wettability of 3D micro/nano silicon structures. Acta Biomater 6(7): 2711-2720.
- Rasala RM, Janorkarc AV, Hirt DE (2010) Poly (lactic acid) modifications. Progress in Polymer Science 35(3): 338-356.
- Roach P, Parker T, Gadegaard N, Alexander MR (2010) Surface strategies for control of neuronal cell adhesion: A review. Surface Science Reports 65(6): 145-173.
- Dehmelt L, Poplawski G, Hwang E, Halpain S (2011) NeuriteQuant: an open source toolkit for high content screens of neuronal morphogenesis. BMC Neurosci 12: 100.
- McCall J, Nicholson L, Weidner N, Blesch A (2012) Optimization of adult sensory neuron electroporation to study mechanisms of neurite growth. Front Mol Neurosci 5: 11.
- Patel A, Li Z, Canete P, Strobl H, Dulin J, et al. (2018) AxonTracer: a novel ImageJ plugin for automated quantification of axon regeneration in spinal cord tissue. BMC Neurosci 19(1): 8.
- Liu K, Lu Y, Lee JK, Samara R, Willenberg R, et al. (2010) PTEN deletion enhances the regenerative ability of adult corticospinal neurons. Nat Neurosci 13(9): 1075-1081.
- Zukor K, Belin S, Wang C, Keelan N, Wang X, et al. (2013) Short hairpin RNA against PTEN enhances regenerative growth of corticospinal tract axons after spinal cord injury. J Neurosci 33(39): 15350-15361.
- Christie KJ, Webber CA, Martinez JA, Singh B, Zochodne DW (2010) PTEN inhibition to facilitate intrinsic regenerative outgrowth of adult peripheral axons. J Neurosci 30(27): 9306-9315.
- Yin H, Kassner M (2016) In Vitro high-throughput RNAi screening to accelerate the process of target identification and drug development. Methods Mol Biol 1470: 137-149.
- Karra D, Dahm R (2010) Transfection techniques for neuronal cells. J Neurosci 30(18): 6171-6177.
- Chen Q, Zhang F, Wang Y, Liu Z, Sun A, et al. (2015) The transcription factor c-Myc suppresses MiR-23b and MiR-27b transcription during fetal distress and increases the sensitivity of neurons to hypoxia-induced apoptosis. PLoS One 10(3): e0120217.
- Dohi E, Tanaka S, Seki T, Miyagi T, Hide I, et al. (2012) Hypoxic stress activates chaperone-mediated autophagy and modulates neuronal cell survival. Neurochem Int 60(4): 431-442.
- Yu Z, Fan D, Gui B, Shi L, Xuan C, et al. (2012) Neurodegeneration-associated TDP-43 interacts with fragile X mental retardation protein (FMRP)/Staufen (STAU1) and regulates SIRT1 expression in neuronal cells. J Biol Chem 287(27): 22560-22572.
- Chih B, Liu P, Chinn Y, Chalouni C, Komuves LG, et al. (2011) A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat Cell Biol 14(1): 61-72.
- Nabzdyk CS, Chun M, Pradhan L, Logerfo FW (2011) High throughput RNAi assay optimization using adherent cell cytometry. J Transl Med 9: 48.
- Zhao M, Yang H, Jiang X, Zhou W, Zhu B, et al. (2008) Lipofectamine RNAiMAX: an efficient siRNA transfection reagent in human embryonic stem cells. Mol Biotechnol 40(1): 19-26.
- Boudes M, Pieraut S, Valmier J, Carroll P, Scamps F (2008) Single-cell electroporation of adult sensory neurons for gene screening with RNA interference mechanism. J Neurosci Methods 170(2): 204-211.
- Tonges L, Lingor P, Egle R, Dietz GPH, Fahr A, et al. (2006) Stearylated octaarginine and artificial virus-like particles for transfection of siRNA into primary rat neurons. RNA 12(7): 1431-1438.
© 2023 Gunnar Poplawski. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)













.bmp)
.jpg)
.png)
.jpg)














.png)

.png)



.png)






