- Submissions

Full Text
Trends in Telemedicine & E-health
A Wiki Based Management System for Clinical Trials Over Semantic Environment
Stella C Christopoulou*
Department of Business and Organization Administration, University of Peloponnese, Greece
*Corresponding author: Stella C Christopoulou, Department of Business and Organization Administration, University of Peloponnese, Antikalamos, Kalamata 24100, Greece
Submission: March 29, 2021; Published: December 01, 2021

ISSN: 2689-2707 Volume 3 Issue 1
Abstract
Despite the growing interest and the challenges associated with collaboration in the process of development and adaptation of knowledge in medical practice on the web there is a lack of experience about the best method to use. This is especially challenging in the area of clinical trials, where healthcare professionals must manage, collaborate, and share a large amount of information from different sources and to work together to acquire the required knowledge in any case. The objective of this preliminary work is to propose a semantic and wiki platform on the web for the management of clinical trials, as an acceptable, feasible, and effective solution.
Keywords: Ontowiki; Wiki; Clinical trials; Semantic web
Abbreviations: OS: Operating System; LDW: Linked Data Wrapper
Introduction
As medical information doubles in volume every three years practitioners are obliged to stay up-to-date with relevant literature and research. Clinical trials are a facet of medical research that constantly increase, while safety directives and regulations demand larger and longer patient participation. Thus it is of critical importance to increase patient enrolment in trials. Along with the increasing efforts for effectiveness, efficiency and safety in healthcare provision, there is a growing interest on best clinical practices, evidence-based medicine and the integration of data quality. The World Health Organization defines a clinical trial as “any research study that prospectively assigns human participants or groups of humans to one or more health-related interventions to evaluate the effects on health outcomes [1].
At the same time domain knowledge has to be refined on the ground of huge piles of
medical data that keep feeding traditional or even semantically enabled databases. Also,
sophisticated data analysis could result to the deduction of common or supplementary results
over relevant or seemingly diverse clinical trials. Clinical trial taxonomy can be based on the
methodology followed by the researchers:
Observational: In an observational study, the investigators study the subjects and
measure their outcomes. The researchers do not actively manage the study.
Interventional:
In an interventional (experimental) study, the researchers administer a particular medicine to the subjects or some other kind of intervention. Usually, they compare the treated subjects to subjects who receive no treatment or standard treatment. Then the researchers measure how the subjects’ health changes. Specifically, in its simplest form, an experimental study to test the effect of a treatment initially the researchers formally state the hypothesis to be tested, select people eligible for the treatment and the sample is divided into two groups. One group (the experimental, or intervention group) is given the intervention while the other (the control group) is not. Finally, outcomes of interest are recorded over time, and the results are compared between the two groups.Another classification of clinical trials is based on their
experimental purpose: There are six types of clinical trials:
a) Diagnostic trials determine better tests or procedures for
diagnosing a particular disease or condition.
b) Natural history studies provide valuable information about
how disease and health progress.
c) Prevention trials look for better ways to prevent a disease
in people who have never had the disease or to prevent the
disease from returning.
d) Quality of life trials (or supportive care trials) explore and
measure ways to improve the comfort and quality of life of
people with a chronic illness.
e) Screening trials test the best way to detect certain diseases or
health conditions.
f) Treatment trials test new treatments, new combinations of
drugs, or new approaches to surgery or radiation therapy [2].
Medical information is dynamically increasing in volume while
relevant knowledge has to be classified and widely provided to
stakeholders. Wikis offer innovative approaches to standardize,
classify, and present clinical research and domain knowledge. The
system proposed in this work aims at providing a wiki based and
semantically enabled system for management of clinical trials. We
mainly investigate four aspects that are of industry and medical
interest:
a) Patient recruitment focuses on applying the necessary
techniques in finding the qualified patients for a trial
b) Trial suitability checks whether or not a clinical trial’s criteria
make it appropriate for a specific patient
c) Data usability verifies that the clinical trial outcomes can be
merged, aggregated or integrated in order to accomplish
systematic reviews or unified results presentation
d) Health Professional enrollment in order facilitate the
collaboration among stakeholders in terms of access
authorization policies on content.
Related work
A wiki can be identified not only as a collaboration platform but a potentially enriching learning and teaching tool, that is easy to use, offering rapid deployment, and facilitation of knowledge transfer [3]. Thus, wiki based collaboration systems have been proposed as a solution to promote co-operation between stakeholders not only in science [4,5] but in industry [6-8] too. Consequently, healthcare sector has attracted relevant research attention. Semantic wikis have been applied for: collaborative authoring of biomedical vocabularies [9] supporting patients daily life [10,11] and clinical protocols modeling [12]. At the same time, considerable research efforts have been focused on the task of formalizing clinical trials and their eligibility criteria. An initial attempt by Ohno-Mechado, Wang, Mar and Boxwala [13] presented an XML-based system that implemented clinical trial protocols as collections of eligibility criteria. Also, a patient assessment module against the criteria was included in order to construct a matching table. Huang, ten Teije and van Harmelen [14], propose a rule-based scheme that is implemented in prolog through reusable components that encompasses three types of knowledge: trial-specific, domain-specific and common. Moreover, in [15] a semantically enabled system is proposed so as to support clinical trials providing automatic recruitment service and trial finding service. Andronikou, Karanastasis, Chondrogiannis, Tserpes, and Varvarigou [16] analyze the data specifications of a clinical trial process plan and focus on the selection of patients according to eligibility criteria. Toward this, a SOA mechanism over a multitude of semantically linked clinical research ontologies is proposed. On the contrary, MacKellar, Schweikert and Chun in [17] introduce a patient-oriented semantic integration approach through RDF.
The objective of this work is to enable patients to search for the proper clinical trial instead of industries look for them. Olasov and Sim in [18] describe RuleEd, a web-based environment for eligibility rules management through appropriate interfaces for interactive concept mappings. Semantic neighbors of a term are displayed according to the UMLS [19] hierarchy. In nowadays there is a continuously developing research in the field of the automatic semantic annotation methods and search of a target corpus using several knowledge resources [20,21]. More specifically in the biomedical domain, there are several clinical text analysis, knowledge extraction and annotation systems that map free biomedical text to standard conceptual form using meta thesaurus like UMLS, SNOMED-CT, NCI-T etc as knowledge sources. Meta Map [21], cTAKES [22], NCBO annotator [23] in BioPortal1 and Semantator [24] are some typical implementations of such systems.
System Design and Functionality
Our proposed system is a knowledge base implemented as a wiki on top of the well-known onto wiki platform. Apart from a typical semantic application, onto wiki2 is an appropriate back office for the new era of semantic and wiki based web information systems. It allows for globalized data manipulation and knowledge management through human-centered user interfaces and extensions. Wiki’s and visualization portals employ onto wiki to provide access in distributed and heterogeneous data or knowledge. It is an Open Source project and Operating System (OS) independent. Thus, it can be deployed over multiple data servers that span from MySQL to virtuoso3. In order to consolidate the wiki nature of our proposed system we incorporated RDF Author that is an onto wiki4 extension for editing distributed content on the Web. This tool contributes to our semantic and wiki environment the features of scalability, evolvement and collaboration.
The semantic library model
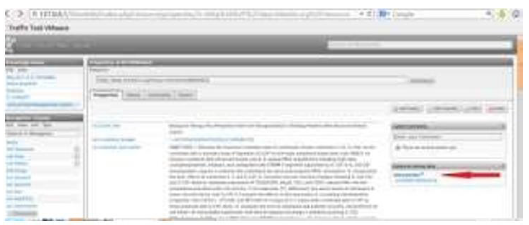
Some of the most basic classes (entities) and properties of wiki based management system for clinical trials’ ontology are described below. Primarily we looked for web data sources about clinical trials. We perceived that there are several open semantic web data sources for clinical trial data. However, LinkedCT [25] (Figure 1) is the first project in the field that publishes linked data on the web improving adaptability and usability. Its database is formed by converting data sources in RDF and identifying semantic links between data.
Figure 1: The NCT00001250 trial as is presented in linkedct.org.

Entity trial
In our knowledge base we created an entity named trial. Subsequently, we linked some indicative trials as instances of the trial entity and their properties from linked CT55 via the Linked Data Wrapper that is an onto wiki extension i.e., a background resources retriever from the data web to the local knowledge base Indicatively (Figure 2), some of the properties retrieved were: brief title, study design, primary completion date, brief summary, number of groups, start date, has expanded access, intervention browse, sponsor group, eligibility, keywords, conditions, locations, interventions, secondary outcomes, references, primary outcomes etc. Many of them are links to other entities that can be included in the knowledge base in a similar way.
Figure 2: The NCT00001250 as imported in Onto Wiki from linkedCT.org via the linked data wrapper.

Entity Trials
However, properties management is a complicated issue that usually requires the involvement of an automatic Semantic Annotation technique to map biomedical text to a standard thesaurus system like UMLS [19]. Nevertheless, the process of the semantic annotation is out of scope in our work. Thus, in the second use case provided (i.e., Plot case 2: Searching using semantic information about eligibility criteria of the trials) we employed an existing library of eligibility criteria that is implemented by Milian, Bucur and Van Harmelen [26]. In their work eligibility criteria acquire structured form and consequently detailed search upon them is possible. For this reason, we selected their library and we imported the relevant entities of this in our system (Figure 3). More specifically a new entity was created, named Trials, as a different one from the existing in our knowledge base i.e., the trial. Then it was linked with the ones imported from the linked CT with the property ‘same As’ (Figure 4).
Figure 3:A part of the NCT00001250 trial that provides semantic information about eligibility criteria as imported in Onto wiki from the existing ontology that provide eligibility criteria of the trials as implemented by Milian, Bucur and Van Harmelen.
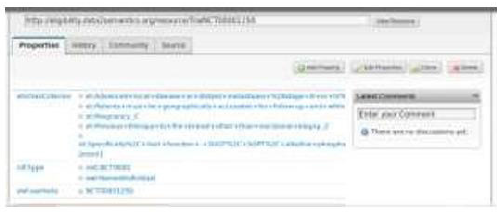
Figure 4:The link between the different instances of a same trial via the property same as.
Entity criterion
The entity Criterion is connected with the Trials entity via the has Criterion property (Object Property). Thus any trial is connected with the appropriate formalized and structured eligibility criteria.
Emerging issues of credibility, confidence and rights:As the issues of confidence and quality of medical information are identified of critical importance during their life cycle, this onto wiki knowledge base does not allow anonymous data modification. Access to medical content follows the approach already proposed in our previous related work [27]. Moreover, this work proposes a relevant schema to monitor and control any content publication. Users, user-groups and access rights are deployed so as to implement a security model that protects against inappropriate or non-authorized content publication.
The Semantic and Wiki Management System for Clinical Trials
User management
As the issues of confidence and quality of medical information are identified as of critical importance during their whole life cycle, our onto wiki knowledge base does not allow anonymous data modification. Access to medical content follows the approach already proposed in [27]. More specifically, in that proposed health ecosystem the users/groups management module enables users to access functionality and data in order to collaborate and cocreate health care content while protecting medical information. Accordingly, in the proposed onto wiki knowledge system herein any health professional (user) must be a member of a research or work group. Users with administrative rights are responsible for monitoring research progress and control published content of any e-care domain. Consequently, any user can manage its own data, link them with other users’ semantic data (located in the system or in the web) or restructure, enrich and share new information generated after processing manually or via specific tools such as semantic annotators.
The satisfaction of the above is achieved by applying FOAF management method [28] that is an important profile supplier as it is currently considered as one of the best populated ontologies. Thus, we created the following FOAF entities in our model to describe groups, users and their relations. More analytically, we created a FOAF person entity that is a healthcare professional, and it is the basic class for describing all users of wiki based management system for clinical trials system. and is subclass of FOAF: Agent. It is defined by some simple FOAF properties (such as FOAF: Name, FOAF: Nick, FOAF: Person’s specialty, etc) and by domain specific properties. The roles of a person are defined by its membership in FOAF: Group. Accordingly, the FOAF group class is used to create basic groups of users. Finally, e-care domain is a class we have created to define the specialty or interest of a user. This data can be used to group users with a similar e-care domain, to recommend additional material or to create an entire domain in a specific healthcare field.
System capabilities
A. The user can manage its own profile and any data with
appropriate access rights, to link them to other users’ semantic data
(located in the system or in the Web) and to restructure, enrich and
share new information and knowledge generated after processing
via relevant tools such as semantic annotators.
B. The system supports online collaboration and co-creation
of medical knowledge between users with the same scientific and
research interests.
Use Case Scenarios
Plot case 1: Transferring and using semantic information from linkedCT
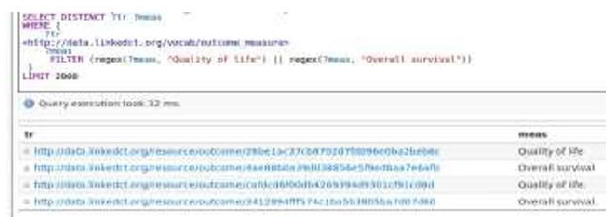
V.D. which is a healthcare professional wants to study the clinical trials related to the quality of life and overall survival of cancer treatment. Initially V.D. logged in the system and created her profile. Consequently, she searched linkedCT by providing the appropriate searching criteria to find specific trials about cancer treatment. Then she imported the trials related to the above mentioned condition via the onto wiki’s Linked Data Wrapper. Then every legal user of the system may search in order to find these trials (Figure 5). Moreover, any specific SPARQL query may be submitted using specific search criteria like brief title, study design, primary completion date, brief summary, number of groups, start date, has expanded access, intervention browse, sponsor group, eligibility, keywords, conditions, locations, interventions, secondary outcomes, references, primary outcomes etc. Finally, V.D. and some other users, with suitable access rights, may manage (create, delete, modify or add) elements of semantic information as entities, properties and instances as an onto wiki user’s domain. For example, the sample SPARQL query (Figure 6) below retrieves trials that have an output measure with a property: outcome measure that contains the phrase either “Quality of life” or “Overall survival”.
SELECT DISTINCT ?tr ?meas WHERE {
?tr
Figure 5:Properties and instances of the imported trials in Onto wiki.

Figure 6:The SPARQL query that retrieves trials which have an output measure with a property: Outcome measure that contains the phrase either “Quality of life” or “Overall survival”.
Plot case 2: Searching using semantic information about eligibility criteria of the trials
We construct a query that requests the trials which have common eligibility criteria with the NCT00002475 trial (Figure 7). The result of this query depicts the trials in descending order with respect to the total number of common eligibility criteria (num Cr) that any trial has with the reference one (NCT00002475 trial), the trial id (trialId) and a brief title (brief_title). The results of the above SPARQL query depict that the trials with Identification (trialId): NCT00014456 and NCT00002475 have four common eligibility criteria. In order to identify them a new SPARQL query is issued. This query illustrates the common eligibility criteria (common criteria) between NCT00014456 and NCT00002475 trials (Figure 8).
Figure 7:A query that requests the trials which have any common eligibility criteria with the NCT00002475 trial.
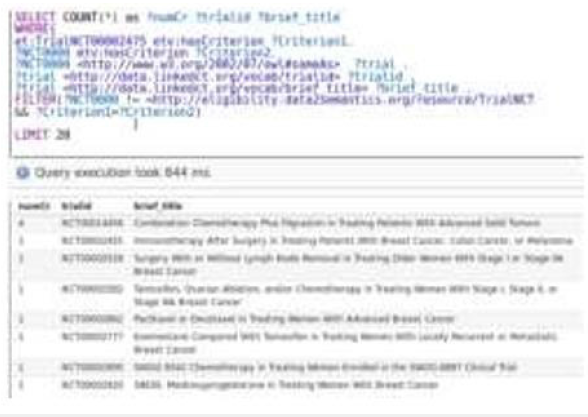
Figure 8:A query that illustrates the common eligibility criteria between NCT00014456 and NCT00002475 trials.

Conclusion and Future Work
Web 2.0 may be one of the most influential technologies in the history of publishing, as old proprietary notions of control and ownership fall away. An expert (i.e., a physician) moderated repository of the knowledge base, in the form of a medical wiki, may be the answer to equal information access in medicine if we have the will to create one [29]. Moreover Web 3.0 directs the systems integration through the development and implementation of semantic rules and the advancement of semantic information. Therefore, in our proposed work that is supported by these technologies all data of the clinical trials may acquire a structured semantic form and consequently thorough search upon them and extraction of medical knowledge is feasible. Also, the management of these data and the co-working is possible for all authenticated users and groups with similar medical interests. Furthermore, we intend to extend our proposed system in accordance with the approach already proposed in [27] across the widespread spectrum of care management by enriching the system mainly with the modules for medical and healthcare guidelines management, the management of a collaborative healthcare community and the relative conformance testing and assessment of the system. Finally, we intend to work towards acquiring a HON code certification [30] for the system so as to verify it as a typical web medical portal.
References
- WHO (2014) World Health Organization. International Clinical Trials Registry Platform (ICTRP).
- (2014) National Institute of Health. NIH Clinical Research Trials and You-Glossary of Common Terms, National Institute of Health.
- Boulos MN, Maramba I, Wheeler S (2006) Wikis, blogs and podcasts: A new generation of Web-based tools for virtual collaborative clinical practice and education. BMC medical education 6(1): 41.
- Wever B, Van KH, Schellens T, Valcke M (2011) Assessing collaboration in a wiki: The reliability of university students' peer assessment. The Internet and Higher Education 14(4): 201-206.
- Silva AR, Rosemann M (2012) Processpedia: An ecological environment for BPM stakeholders' collaboration. Business Process Management Journal 18(1): 20-42.
- Holtzblatt LJ, Damianos LE, Weiss D (2010) Factors impeding wiki use in the enterprise: A case study. In CHI'10 Extended Abstracts on Human Factors in Computing Systems, pp. 4661-4676.
- Blohm I, Bretschneider U, Leimeister JM, Krcmar H (2011) Does collaboration among participants lead to better ideas in IT-based idea competitions? An empirical investigation. International Journal of Networking and Virtual Organisations 9(2): 106-122.
- Grudin J, Poole ES (2010) Wikis at work: Success factors and challenges for sustainability of enterprise wikis. Proceedings of the 6th international symposium on Wikis and open collaboration.
- He S, Nachimuthu SK, Shakib SC, Lau LM (2009) Collaborative authoring of biomedical terminologies using a semantic wiki. In AMIA Annual Symposium Proceedings, pp. 234-238.
- Archambault PM (2011) Wikibuild: A new application to support patient and health care professional involvement in the development of patient support tools. Journal of medical Internet research 13(4): 114.
- Gupta S, Wan FT, Newton D, Bhattacharyya OK, Chignell MH, et al. (2011) Wiki build: A new online collaboration process for multistakeholder tool development and consensus building. Journal of Medical Internet Research 13(4): 108.
- Eccher C, Ferro A, Seyfang A, Rospocher M, Miksch S (2009) Modeling clinical protocols using semantic Media wiki: The case of the oncocure project. Knowledge management for health care procedures, pp. 42-54.
- Ohno-Machado L, Wang SJ, Mar P, Boxwala AA (1999) Decision support for clinical trial eligibility determination in breast cancer. Proceedings of the AMIA symposium, pp. 340-344.
- Huang Z, Teije AT, Harmelen F (2013) Rule-based formalization of eligibility criteria for clinical trials. Conference on Artificial Intelligence in Medicine in Europe, pp. 38-47.
- Huang Z, Teije A, Harmelen F (2013) SemanticCT: A Semantically-Enabled System for Clinical Trials. Springer, pp. 11-25.
- Andronikou V, Karanastasis E, Chondrogiannis E, Tserpes K, Varvarigou T (2010) Semantically-enabled intelligent patient recruitment in clinical trials. 2010 International Conference on P2P, Parallel, Grid, Cloud and Internet Computing.
- MacKellar B, Schweikert C, Chun S (2013) Patient-oriented clinical trials search through semantic integration of linked open data. 2013 IEEE 12th International Conference on Cognitive Informatics and Cognitive Computing.
- Olasov B, Sim I (2006) RuleEd, a web-based semantic network interface for constructing and revising computable eligibility rules. AMIA Annual Symposium Proceedings.
- Bodenreider O (2004) The Unified Medical Language System (UMLS): Integrating biomedical Nucleic acids research 32(1): 267-270.
- Berlanga R, Nebot V, Pérez M (2014) Tailored semantic annotation for semantic search. Journal of Web Semantics 30: 69-81.
- Aronson AR, Lang FM (2010) An overview of metamap: Historical perspective and recent Journal of the American Medical Informatics Association 17(3): 229-236.
- Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, et al. (2010) Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): Architecture, component evaluation and applications. Journal of the American Medical Informatics Association 17(5): 507-513.
- Noy N, Shah NH, Whetzel PL, Dai B, Dorf M, et al. (2009) BioPortal: Ontologies and integrated data resources at the click of a mouse. Nucleic Acids Research.
- Tao C, Song D, Sharma D, Chute CG (2013) Semantator: Semantic annotator for converting biomedical text to linked data. Journal of Biomedical Informatics 46(5): 882-893.
- Hassanzadeh O, Kementsietsidis A, Lim L, Miller R, Wang M (2009) LinkedCT: A linked data space for clinical trials. p. 1-5.
- Milian K, Bucur A, Harmelen F (2012) Building a library of eligibility criteria to support design of clinical trials. Knowledge Engineering and Knowledge Management, pp. 327-336.
- Christopoulou S, Kotsilieris T, Dimopoulou N (2013) A web tool for an Open and Linked Health Ecosystem. In E-Health and Bioengineering Conference (EHB).
- Dan B, Libby M (2014) FOAF Vocabulary Specification 0.99. Namespace Document 14, Paddington Edition.
- Stiglitz JE (2006) Scrooge and intellectual property rights. BMJ 333(7582): 1279-1280.
- Boyer C, Selby M, Scherrer JR, Appel R (1998) The health on the net code of conduct for medical and health websites. Computers in Biology and Medicine 28(5): 603-610.
© 2021 Stella C Christopoulou. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






