- Submissions

Full Text
Significances of Bioengineering & Biosciences
A Review on MiRNA Biogenesis
Aashi Fazeela M*, Jaya Joshi, Ashish Verma and Ankit Goyal
Department of Oral Pathology & Microbiology, Government College of Dentistry, Indore, Madhya Pradesh, India, 452001
*Corresponding author: Aashi Fazeela M, Department of Oral Pathology & Microbiology, Government College of Dentistry, 1-Sardar Patel Marg, Indore, Madhya Pradesh, India, 452001 Email ID: aashifazeela@gmail.com
Submission: March 16, 2021 Published: April 26, 2021

ISSN 2637-8078Volume4 Issue5
Abstract
Micro RNAs are single stranded non-coding RNAs that function to regulate the mRNA and thereby proteins. It functions as an important epigenetic biomarker. The multiverse of miRNA is highly complicated one which is governed by numerous protein miRNA interactions which causes alteration in its genesis as well as regulation. To understand the importance of miRNA in our genome, it is essential to have a clear knowledge of the various pathways leading to its formation. This pathway occurs in different levels and include the interplay between many enzymes such as Drosha, Dicer. Following the genesis, matured miRNA becomes active and complementarily binds with mRNA to cause its silencing/exaggeration. This genesis of miRNA as well as its interaction is under the influence of many regulatory proteins.
Keywords: MiRNA biogenesis; Canonical pathway; Non-canonical pathway; Regulation; mRNA silencing; Enhanced translation
Abbreviations: piRNA: Piwi Interacting RNA; miRNA: MicroRNA; DGCR8: Digeorge Syndrome Critical Gene Region in Gene 8; EXP5: Exportin 5; TRBP: Trans-activation Responsible DNA Binding Protein; RISC: RNA Induced Silencing Complex; UTR: Untranslated Region; shRNA: Short Hairpin RNA
Introduction
In eukaryotes, multiple small RNAs have evolved to control abnormal expression of genetic information and proteins. Those RNAs that are of 20-30 nucleotide in length and associated with argonate family proteins are categorized as small RNAs. In animals, three types of small RNAs are majorly known and they include MicroRNA, siRNA and piwi interacting RNA (piRNA). MicroRNAs are one of the major/dominating type of single stranded non-coding small RNAs of somatic cells composed of around 22 nucleotides [1]. More than 50% of human genes are regulated by micro RNAs. It influences basic cellular processes like development, differentiation, proliferation, apoptosis, and regulation of cell cycle. miRNA pleiotropically regulates the mRNA translation and decay thereby significantly acting as a post transcriptional regulator [2]. miRNA genomics received its first milestone when a small RNA, lin-4A was discovered in 1993 in nematodes using genetic screening technique by Victor Ambros and colleagues. As its functions was unknown, historically tit was referred to as junk DNA. Later, in the same year, the regulatory role of lin4A was discovered [3-5]. Yet, the era of miRNA genomics only began in 2000 with the discovery of let-7 RNA in Caenorhabditis elegans, followed by in humans [6]. Today more than 1800 miRNAs has been discovered. Owing to the recent technological advancements such as high-throughput sequencing technologies, computational and bioinformatics prediction methods, the research on miRNA is progressing at a lightning speed [5]. But still, the specific biological function of these identified molecules are unknown. Studies comparing the miRNA expression profiles definitely provides significant information regarding its function and regulation in normal and diseased conditions. This sheds light onto the possible role of miRNA in pathogenesis of multiple diseases including cancer. To have an in-depth concept, it is essential to understand the basics of miRNA in terms of its biogenesis and regulation. Through this article, an attempt has been made to summarize the formation, effects and regulation of this biomolecule, providing an insight into the beautiful world of miRNAs.
MicroRNA Biogenesis
Synthesis of mature miRNA is a complicated pathway that involves multi-step processing of primary RNA. miRNA biogenesis starts with the action of RNA polymerase II/III on the precursor and is classified into Canonical and Noncanonical pathways.
Results
Canonical pathway of miRNA biogenesis
This is the most dominant pathway for miRNA synthesis which is getting regulated by numerous enzymes in a complex manner. It is initiated in nucleus and is completed in the cytoplasm. Biogenesis is better understood if described in a sequential order as given below:
miRNA precursor genes: miRNA precursor genes are those genomic regions that codes for miRNA biogenesis. These are commonly found within different areas of genome. In most of the scenario miRNA precursor genes are non-coding genes i.e., they do not code for any other mRNA/protein. In other words, their sole purpose is to generate miRNA [6]. Based on its location within the genome, it can be either intergenic (between two genes) or intragenic (i.e., within a gene) (Figures 1A & 1B). According to early researchers, majority of miRNAs are located in intergenic region whereas only few were found in intronic regions, which is one of the most common intragenic region [7-9]. The transcriptional units and regulation of biogenesis of miRNA alters with the change in the precursor gene location. Intronic miRNAs, located within the genome share the same promoter along with the encoded target genes. Here, Intronic sequences are spliced out of the transcript of encoded genes during post transcriptional modification. These intronic sequences are further undergoes modifications and forms mature miRNAs. On the other hand, Intergenic regions act as an independent unit and has its own promoter along with other regulatory system. miRNA genes are mainly transcribed by the enzyme RNA polymerase II but RNA polymerase III also participate in this process as it transcribes a small group mainly the one containing Alu repeats [10,11].
Figure 1A: Intergenic genome showing miRNA between two genes.
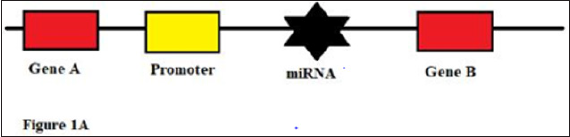
Figure 1B: Intronic miRNA located in the intronic region of the genome.

Nuclear processing: Poly II/Poly III enzymes mediates transcription of miRNA genes and produces primary miRNA (primiRNA). These pri-miRNAs are several kilobases long, contains local stem loop structures and single stranded RNA segments at both the 5’ and 3’ side (Figure 2). Hence it requires further processing. All animal pri-miRNAs are processed first within the nucleus by a microprocessor complex formed of an enzyme Drosha along with a cofactor Digeorge Syndrome Critical Gene Region in Gene 8(DGCR8). This complex cleaves the pri-miRNA at the stem of hairpin structure leading to release of a structure containing 60-70 nucleotide(nt). This structure is known as precursor miRNA (premiRNA) [12-14].
Figure 2: Intronic miRNA located in the intronic region of the genome.

Transportation to cytoplasm: The maturation of miRNA occurs in the cytoplasm for which the pre miRNA formed in nucleus needs to be transported. This is done through large protein channels embedded in nuclear membrane known as nuclear pore complex [15]. The transport is further mediated by Ran GTP dependent nuclear transport receptor Exportin 5(EXP5) complex (Figure 2). The crystal structure of this complex shows the presence of many baseball mitt like structures onto which pre-miRNA stem can be placed. This enables the interaction of pre-miRNA stem and the EXP5 Complex. Also, a basic tunnel like structure present at the bottom of the mitt like structure can interact with 2 nucleotide long 3’ structure of pre-miRNA [16]. These interactions bind pre-miRNA to EXP5 and thereby facilitate the transportation from the nucleus through nuclear pore into the cytoplasm. Subsequently, hydrolysis of GTP releases the pre-miRNA from the complex [5].
Cytoplasmic processing: The pre-miRNA released into the cytoplasm by EXP5 is processed by a highly specific enzyme known as Dicer. The human Dicer is also accompanied by two proteins namely Trans-activation Responsible DNA Binding Protein (TRBP) and protein kinase, interferon inducible double stranded RNA dependent activator (PRKA also called as PACT). Dicer identifies and cleaves 22 nucleotide near to the terminal loop of the transported pre-miRNA. Hence, a 22 nucleotide miRNA duplex is formed (Figure 2) [5]. This 22nucleotide miRNA duplex, containing one mature miRNA guide strand (miRNA) and one passenger strand(miRNA*) gets incorporated into Ago family protein. Chaperone machinery HSP70/ HSP90 aids in this process. All these together i.e. miRNA duplex, dicer and Ago proteins, form an effector complex [17- 19]. Dicer, its associated proteins and Ago family are responsible for pre-miRNA processing and they also lead to formation of RNA Induced Silencing Complex (RISC) assembly [5,20].
RISC assembly: RISC is a complex composed of miRNA, dicer, its associated proteins and Ago. MiRNA within the RISC guides the complex to bind to mRNA and regulate its function. Information regarding the exact composition of RISC is unknown but it is clear that it contains an essential protein called argonate. Humans have 8 classes of RISC complex based on its protein composition that is chiefly centered around 4 argonate proteins [21,22]. Out of all these, only AGO2 has slicing activity that helps it to catalyze the mRNA cleavage [14]. The resultant miRNA duplex(miRNA/miRNA*) produced as a result of dicer cleavage binds to Ago Protein. miRNA* of the duplex is degraded by one of the many possible mechanism given below.
a. If the miRNA duplex has complementarity around the
central region; it is cleaved by AGO2.This commonly occurs in
the case of siRNA.
b. But most miRNA duplexes lack this central
complementarity and hence rely on unwinding and cleavage by
an unknown helicase for its degradation [6].
RISC with incorporated miRNA is referred to as miRNA
containing nucleoprotein complex (miRNP) [23]. MiRNA within the
RISC guides the complex to bind to mRNA and regulate its function. Formation of RISC also offers stability to both the miRNA and
the Ago protein which otherwise may get destabilized via Tudor-
SN-mediated endonucleolytic decay or target directed miRNA
destabilization (it involves trimming and 3’ end tailing) [24].
Action on mRNA: The action of miRNA on mRNA is partly
based on the complementarity between them. Nucleotide 2-7 of
miRNA is termed as the seed region [25]. This region as well as the
3’ end is important for the association between miRNA and mRNA.
Centrally matched targets have also been identified. If central
complementarity exists, then the mRNA target can be cleaved via
the endonuclease activity of AGO2 present within the RISC. But
this does not happen in the case of humans, as they lack central
complementarity [5,23]. miRNA can induce target repression with
help of TNRC6 proteins that shortens the poly(A) tails of mRNA
there by repressing its translation. It can also destabilise the mRNA
[24].
Non-canonical miRNA biogenesis pathway
Many non-canonical/alternative miRNA biogenesis pathways have been identified. This pathway uses different combinations of the canonical proteins such as Drosha, exportin 5, Dicer and AGO2 i.e., it primarily consists of Drosha/DGCR8-independent and Dicer independent pathways [5].
The pre-miRNA produced due to Drosha independent pathway resembles substrates for DICER. Some examples include:
a. Mirtrons: When mRNA undergoes splicing, a microRNA called as mirtrons are created from intronic sequences
b. 7-methylguanosine (m7G)-capped pre-miRNA: These nascent RNAs are directly exported to the cytoplasm through exportin 1 without the need for Drosha cleavage.
Dicer independent miRNA: Dicer independent miRNA are processed by Drosha from endogenous short hairpin RNA (shRNA) transcripts. Dicer does not identify them as substrate because of their insufficient length. Due to this these pre-miRNA uses AGO2 for their maturation [26,27].
Regulation of miRNA Biogenesis
Most of the studies done in the previous periods focused primely on the transcriptional regulation of the miRNA genes. For example, it is proven that cardiomyocyte transcription factor networks transcriptionally regulate the cardiac-specific miRNA families miR- 1 and miR-133 [28,29]. In the same way, intronic miRNA genes are regulated by the same transcription factor network that manages the coding mRNA [6]. Apart from transcriptional regulation, miRNA biogenesis can also be post transcriptionally regulated [30]. This can be explained using an example of “regulation of let-7 family during differentiation of embryonic cells”. Mature let-7 is normally undetectable in stem cells. But during differentiation let-7 family becomes so highly expressed that it is one of the most abundant miRNA family in differentiated cells [31]. This increased expression happens without any significant increase in the host gene transcription. In other words, it can be stated that let-7 maturation is blocked in embryonic cells. This was found to be done by Lin28. This particular protein:
a) Binds to pri-miRNA and inhibits cleavage by Drosha
b) Binds to pre-miRNA and inhibits cleavage by Dicer
c) Adds oligouridine tail to let-7 precursors and marks them
for degradation.
During differentiation, the expression of Lin28 is shut down due to which a rapid increase in maturation of lin-7 occurs [6,32,33]. To re-evaluate importance of Drosha, Dicer, XPO5, TRBP, and PACT in miRNA biogenesis, several studies using knock out cells have been performed recently. In studies conducted in Drosha knock out cells, production of miRNA was observed to be completely abolished but in Dicer knock out cells only 80% reduction in biosynthesis were observed [34]. In other hand knock out models of XPO5 showed only minimal depletion suggesting the presence of an alternative transport mechanism that is independent of XPO5 [35]. A high throughput analysis of pri-miRNA revealed that many sequence features of it are important for efficient pri-miRNA processing. Some of the features include: a) UG motif at the base of hairpin loop of pri-miRNA. b) UGU motif at the apical loop, a CNNC motif near the Drosha processing site [36]. These structural characteristics causes the efficient interaction between pri-miRNA and Dicer/ DGCR8 complex.
Mechanism of Gene Regulation by miRNA
Most of the discovered miRNAs bind to the 3’ UTR region of mRNA. But some of the miRNAs are also seen to be bind to 5’ UTR region, coding regions as well as promotor regions of mRNA [37,38]. It is observed through various studies that when a miRNA is bound to 5’UTR regions and coding regions, it has a silencing action whereas when bound to promoter region it induces transcription of mRNA [39].
Silencing of gene via RISC assembly
miRNAs present within the miRISC guides it to identify mRNA and to alter its expression. There is a downregulation of mRNA expression either via translational repression or via mRNA cleavage [33]. This is dependent on the degree and nature of complementarity between the guiding miRNA and mRNA. When there is a near perfect base pairing between the strands; AGO2 cleavage is favoured [14,40] whereas translational inhibition requires multiple complementary sites with only limited base pairing. mRNA cleavage and translational repression are also referred to as slicer dependent and slicer independent silencing respectively. Though both of these mechanisms ultimately lead to downregulation of gene expression, the significant difference between the effects is its reversibility. Translational inhibition is reversible because stable mRNA can be translated following elimination of translational repression whereas slicer/AGO2 dependent cleavage is irreversible [14].
Micro RNA mediated enhanced translation
Contrary to the traditional role of miRNA in post translational repression, in some cases it can cause enhanced translational activities. Joan and Sietz et al observed the property of increased quiescence of cells in stressful conditions i.e., cells under challenging situations exit from their cell cycle and enter a resting phase. They discovered that miRNA has an important role in it [41]. Steitz [42] through their studies has observed that AGO2 protein and Fragile X Metal Retardation Related Protein 1 (FXR1) causes increased translation. Both these proteins binds to the 3’ Untranslated Region (UTR) of TNF alpha mRNA. After computational research, they found that miRNA 369-3 forms direct base pairing with the target and this process is mandatory for the translational upregulation seen in stressful conditions such as serum starvation [42].
Conclusion
The current proteomic era identifies miRNA as a key player in the pathogenesis and progression of multiple diseases. decade of strong basic research has been performed. Now, it is the time to link this core concept of miRNA with many human diseases, to identify miRNA role in multiple diseases, to identify and establish specific miRNA biomarkers that aid in early detection of a disease. miRNA diagnostics and therapeutics can be developed with confidence and later commercialized to be of effective use with the aim of prevention, prognosis, and early diagnosis. A Core concept of miRNA in various human diseases with the strong basic research of this novel biomolecule in the identification, initiation and progression of the disease is the need of the hour.
References
- Ha M, Kim VN (2014) Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15(8): 509-524.
- Solomon MC, Radhakrishnan RA (2020) MicroRNA's - The vibrant performers in the oral cancer scenario. Jpn Dent Sci Rev 56(1): 85-89.
- Lee RC, Feinbaum RL, Ambros V (1993) The Caenorhabditis elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75(5): 843-854.
- Wightman B, Ha I, Ruvkun G (1993) Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in Caenorhabditis elegans. Cell 75(5): 855-862.
- Wahid F, Shehzad A, Khan T, Kim YY (2010) MicroRNAs: synthesis, mechanism, function, and recent clinical trials. Biochim Biophys Acta 1803(11): 1231-1243.
- Hammond SM (2015) An overview of microRNAs. Adv Drug Deliv Rev 87: 3-14.
- Rodriguez A, Griffiths Jones S, Ashurst JL, Bradley A (2004) Identification of mammalian microRNA host genes and transcription units. Genome Res 14(10): 1902-1910.
- Lau NC, Lim LP, Weinstein EG, Bartel DP (2001) An abundant class of tiny RNAs with probable regulatory roles in caenorhabditis elegans. Science 294(5543): 858-862.
- Lagos Quintana M, Rauhut R, Lendeckel W, Tuschl T (2001) Identification of novel genes coding for small expressed RNAs. Science 294(5543): 853-858.
- Cai X, Hagedorn CH, Cullen BR (2004) Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10(12): 1957-1966.
- Lee Y, Kim M, Han J, Yeom KH, Lee S, et al. (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23(20): 4051-4060.
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN (2002) MicroRNA maturation: stepwise processing and subcellular localization. EMBO J 21(17): 4663-4670.
- Zeng Y, Cullen BR (2003) Sequence requirements for micro RNA processing and function in human cells. RNA 9(1): 112-123.
- O'Brien J, Hayder H, Zayed Y, Peng C (2018) Overview of microrna biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne) 9: 402.
- Nakielny S, Dreyfuss G (1999) Transport of proteins and RNAs in and out of the nucleus. Cell 99(7): 677-690.
- Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, et al. (2009) A high-resolution structure of the pre-microRNA nuclear export machinery. Science 326(5957): 1275-1279.
- Kim VN, Han J, Siomi MC (2009) Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10(2): 126-139.
- Inui M, Martello G, Piccolo S (2010) MicroRNA control of signal transduction. Nat Rev Mol Cell Biol 11(4): 252-263.
- Hutvagner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, et al. (2001) A cellular function for the RNA interference enzyme dicer in the maturation of the let 7 small temporal RNA. Science 293(5531): 834-838.
- Khvorova A, Reynolds A, Jayasena SD (2003) Functional siRNAs and miRNAs exhibit strand bias. Cell 115(2): 209-216.
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ (2001) Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293(5532): 1146-1150.
- Grishok A, Sinskey JL, Sharp PA (2005) Transcriptional silencing of a transgene by RNAi in the soma of Caenorhabditis elegans. Genes Dev 19(6): 683-696.
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, et al. (2005) TRBP recruits the Dicer complex to AgO2 for microRNA processing and gene silencing. Nature 436(7051): 740-744.
- Matsuyama H, Suzuki HI (2019) Systems and synthetic microRNA biology: From biogenesis to disease pathogenesis. Int J Mol Sci 21(1): 132.
- Grimson A, Farh KK, Johnston WK, Garrett Engele P, Lim LP, et al. (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27(1): 91-105.
- Ruby JG, Jan CH, Bartel DP (2007) Intronic microRNA precursors that bypass drosha processing. Nature 448(7149): 83-86.
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R (2008) Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor independent, dicer-dependent small RNAs. Genes Dev 22(20): 2773-2285.
- Baskerville S, Bartel DP (2005) Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 11(3): 241-247.
- Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, et al. (2007) An E2F/miR-20a autoregulatory feedback loop. J Biol Chem 282(4): 2135-2143.
- Thomson JM, Newman M, Parker JS, Morin Kensicki EM, Wright T, et al. (2006) Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev 20(16): 2202-2207.
- Newman MA, Thomson JM, Hammond SM (2008) Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14(8): 1539-1549.
- Viswanathan SR, Daley GQ, Gregory RI (2008) Selective blockade of microRNA processing by Lin28. Science 320(5872): 97-100.
- Kawamata T, Tomari Y (2010) Making RISC. Trends Biochem Sci 35(7): 368-376.
- Kim YK, Kim B, Kim VN (2016) Re-evaluation of the roles of DROSHA, export in 5, and DICER in microRNA biogenesis. Proc Natl Acad Sci USA 113(13): E1881-E1889.
- Kim Y, Yeo J, Lee JH, Cho J, Seo D, et al. (2014) Deletion of human tarbp2 reveals cellular microRNA targets and cell-cycle function of TRBP. Cell Rep 9(3): 1061-1074.
- Fang W, Bartel DP (2015) The menu of features that define primary MicroRNAs and enable de novo design of MicroRNA Genes. Mol Cell 60(1): 131-145.
- Xu W, San Lucas A, Wang Z, Liu Y (2014) Identifying microRNA targets in different gene regions. BMC Bioinformatics 15(7): S4.
- Zhang J, Zhou W, Liu Y, Liu T, Li C, et al. (2018) Oncogenic role of microRNA 532-5p in human colorectal cancer via targeting of the 5′ UTR of RUNX3. Oncol Lett 15(5): 7215-7220.
- Dharap A, Pokrzywa C, Murali S, Pandi G, Vemuganti R (2013) MicroRNA miR 324-3p induces promoter-mediated expression of RelA gene. PLoS ONE 8(11): e79467.
- Jo MH, Shin S, Jung SR, Kim E, Song JJ, et al. (2015) Human argonaute 2 has diverse reaction pathways on target RNAs. Mol Cell 59(1): 117-124.
- Rusk N (2008) When microRNAs activate translation. Nat Methods 5: 122-123.
- Vasudevan S, Tong Y, Steitz JA (2007) Switching from repression to activation: microRNAs can up-regulate translation. Science 318(5858): 1931-1934.
© 2021 © Aashi Fazeela M. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






