- Submissions

Full Text
Significances of Bioengineering & Biosciences
Exploration of Organic Compound Degradation Capability of Indigenous Soil Bacteriaand their Consortium from Oil Refinery of Assam
Eram Ayaz* and Ragini Gothalwal
Department of Biotechnology and Bioinformatic Centre, Barkatullah University, India
*Corresponding author: Eram Ayaz, Department of Biotechnology and Bioinformatic Centre, BarkatullahUniversity, Bhopal (MP), India
Submission: November 25, 2020 Published: December 22, 2020

ISSN 2637-8078Volume4 Issue3
Abstract
Present study was focused on the biodegradation capability of indigenous bacterial culture and their consortium. On the basis of GC-FID, Consortium Qs15 was found to be potent consortium for the degradation of petrol, toluene, xylene, diesel, benzene and hexane. Gene which is responsible for the degradation is present on the plasmid. The amplification and sequencing of the 16S rRNA gene showed, Consortium Qs15 comprised of two Bacillus strains namely, Bacillus sp E14 and Bacillus sp E87. Experimental data reveals that Qs15 proven to be better option to eradicate wide range of hydrocarbon via bioremediation technique.
Keywords: Hydrocarbon; Intrinsic; Indigenous; Consortium
Introduction
Spillage and discharges of hydrocarbons occurred due to storage, transportation, refining and processing of petroleum compound, these activities include leakage from oil pipelines, storage tanks, tanker leakage accidents, oil well waxing, overhauls of refineries and production of petrochemical equipment. Petroleum hydrocarbon is a crude mixture of alkane, cyclic alkane, alkene, and aromatic hydrocarbon, which are found to be highly toxic and carcinogenic in nature [1]. Many normal and extreme bacterial species have been isolated and utilized as biodegraders for dealing with petroleum hydrocarbons for bioremediation. This is also indicating that the joint action of multiple functional bacteria requires to achieve the best environmental purification effect for the remediation of petroleum hydrocarbon contamination [2]. The advantages of microbial communities are presented because there are a variety of catabolic genes in a bacterial consortium, and the synergistic effects of these genes are beneficial to achieve the purification of pollutants [3]. In their finding, it was concluded that due to the abundance of alk B and nah genes responsible for the catabolism of hydrocarbons consortium show the maximum degradation as compared to individual isolates.
Materials and Methods
Isolation of bacteria
The bacteria were isolated by using standard protocol of Xiao and Zhang (2011) in mineral salt medium (MSM) supplemented with 1% of each hydrocarbon such as, diesel, petrol, xylene, benzene, toluene and hexane) [4]. The flask was incubated at 37 °C on a rotatory shaker at 100rpm for 48hrs. Three successive sub-culturing were done with 1% hydrocarbon. 0.1ml of culture was plated and incubated at 37 °C for 48hrs from the third sub-culturing. After over incubation times pure colonies were isolated by using single colony isolation methods [5].
Screening of isolates
Exponentially growing and pure bacterial culture (24hrs old) were inoculated in mineral salts medium with 0.006%, 0.008% and 0.01% concentrations of selected hydrocarbons (benzene, diesel, hexane, petrol, toluene and xylene). They were kept in a shaker at 200rpm at 35 °C for a period of 96hrs. The growth was monitored at regular interval of 24hrs up to 96hrs through culture densities, measuring the absorption spectrophotometrically at 595nm [6].
Plasmid curing
To check whether the oil degrading gene are plasmid encoded or chromosomal encoded modified method of Trevors [7] was used for the curing of plasmid. The bacterial isolates Gh1 and Gh15 were cultured in 50ml MSM containing ethidium bromide (20mgml-1) and incubated at 37 °C for 24hrs [7]. Thereafter, the broth was agitated to homogenize the content and 1ml of cultures was sub cultured 50ml MSM medium containing 100ppm of each hydrocarbon individually. The flasks were then incubated at 37 °C for 24hrs and the optical density was taken. Isolates that failed to grow on MSM medium were considered as cured.
Consortium preparation
A loop full of overnight grown culture was used to inoculate 25ml sterile mineral salt medium. The flasks were kept in a shaker at 200rpm for 24hrs at 35 °C. Equal volumes of culture broth from the selected isolates were used to prepare the bacterial consortium. For the preparation of consortium only those strain were selected which have the capability to degrade all the selected hydrocarbons. Selection criteria of hydrocarbon is based on diverse range of hydrocarbon such as aromatic, aliphatic and complex hydrocarbon due to which selected consortium will be used as potent agent for degradation of wide spectrum of hydrocarbons. This bacterial consortium (1ml) inoculated in 50ml mineral salt medium with 0.01% of different hydrocarbon. The growth was monitored through culture densities, measuring the absorption at 595nm, spectrophotometrically Boboye et al. [5] at regular interval of 24hrs [5]. Quantitative estimation of protein was also performed [8]. The nomenclature of consortium have been illustrated which are as follows (Table 1).
Table 1: The nomenclature of consortium.
Identification of the Members of Selected Consortium
The molecular identification of bacterial isolates in which the genomic DNA was extracted using standard protocol [9]. The16S rDNA was polymerase chain reaction (PCR) amplified using universal primer pair, 968F (5’-AGAGTTTGATCMTGG-3’) and 1541R (5′-TACCTTGTTACGACTT 3′) (White et al. 1990). PCR was performed with final volume of 50μl in thermal cycler (Mastercycler Nexus gradient, Eppendorf, Germany) with a final concentration of 1X standard buffer, 1.5mmoll-1 MgCl2, 0.25μmoll-1 each primer, 0.25mmoll-1 dNTPs and 1.0U Taq DNA polymerase (Sigma-Aldrich, USA) and 25ng of template DNA. The PCR reaction conditions consisted of initial denaturation at 95 °C for 7 minutes, followed by 35 cycles of denaturation at 94 °C for 1min, annealing at 51 °C for 1min, extension for 1min at 72 °C, and a final extension for 10min at 72 °C for PCR products were analyzed on 0.8% agarose gel and visualized under UV transilluminator. From PCR products, unincorporated PCR primers and dNTPs were removed by using Montage PCR clean up kit (Millipore). Sequencing of PCR product was done in 3730XL automated DNA sequencing system (Applied Bio Systems, USA). 16S rDNA consensus sequence was used for Basic Local Alignment Search Tool (BLAST) analysis against nr database in the National Centre for Biotechnology Information (NCBI) GenBank. Aligning of sequence data were done using Clustal W (Thompson et al. 1994) and phylogenetic relationship among the strains were determined by neighbor-joining method using MEGA 6 software [10].
Gas Chromatographic Analysis of Extracted Hydrocarbons by Individual Isolates
The hydrocarbons (Benzene, hexane, toluene and xylene) were extracted from liquid cultures with acetone. The extracts were then measured by direct injection into a Gas chromatograph equipped with an FID and 30mx0.25umx250um (diameter) fused silica capillary column (PE-FFA8). Flow rate of nitrogen was 6psi and the sample size was 5μl. The injection and the detector were maintained at 230 °C, and the oven temperature was programmed to rise from 10 °C/min to 210 °C/min and to hold at 210 °C for 10min. For the analysis of diesel and petrol BP-5 column was used with the following dimension: 15mx0.35umx2.50um. Nitrogen was the carrier gas at a flow rate of 6psi. The injection port was set at 250 0C. The oven temperature was programmed from 10 0C/min to 280 0C, holding the temperature for 10min [11].
Results and Discussion
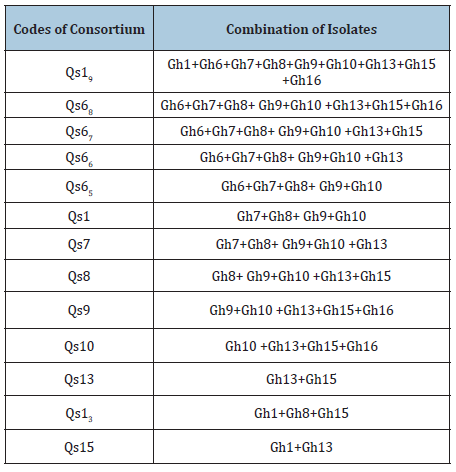
On the basis of turbidity, it was concluded only 10 isolates have the ability to utilize hydrocarbons as carbon source (Figure 1) Gh1 and Gh15 showed the better efficiency towards the higher concentration of selected hydrocarbons. To check whether this hydrocarbon degrading ability is plasmid encoded or not, plasmid curing process was done (Figures 2 & 3). Curing experiments demonstrated that cured isolates was unable to grew in hydrocarbons supplemented media. Therefore, biodegradation potential shown by the strain is plasmid mediated. Catabolic pathways involved in aromatic hydrocarbon degradation routes are located on large plasmids in most of the cases although degradative genes can be located on either chromosome or plasmid or on both [12]. The TOL plasmid pWWO of Pseudomonas putida mt-2 is a 117-kilobase (kb) catabolic plasmid which has ability to encodes all enzymes necessary for bacterial utilization of toluene (Tol), m- and p-xylene (Xyl), 3-ethyltoluene, and 1,2,4-trimethylbenzene and carboxylic acid derivatives, via a meta cleavage pathway [13].
Figure 1:Growth of selected morphtypes in presence of tested hydrocarbons at 100ppm.
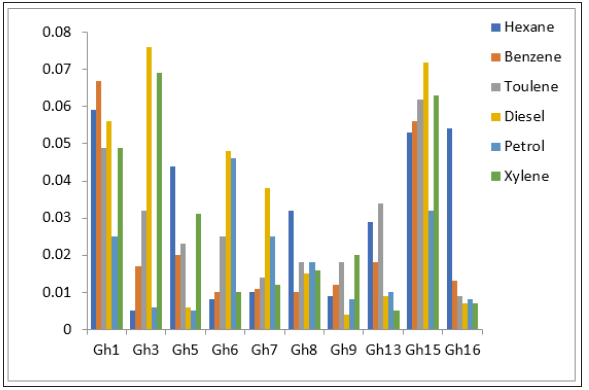
Figure 2:Growth pattern of Gh1 with cured and a. uncured plasmid, b. In the presence of selected hydrocarbons.
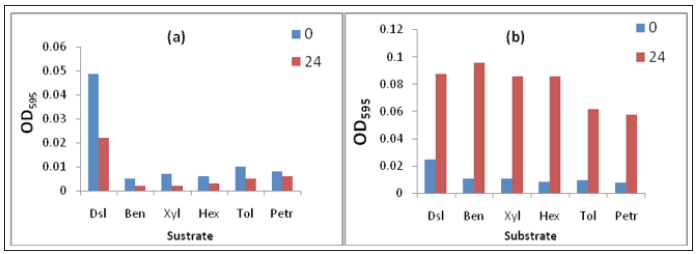
Figure 3:Growth pattern of Gh15 with cured a. uncured plasmid and b. In the presence of selected hydrocarbons.
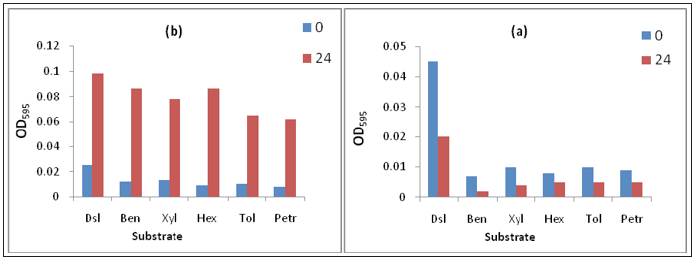
Figure 4:Growth pattern of consortium Qs15 on different substrate (100ppm of each).
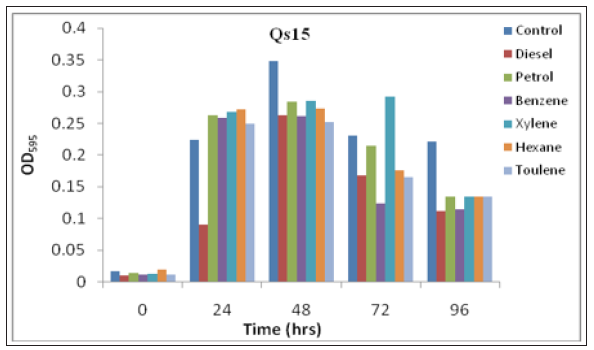
These two isolates selected for further study by the formation of consortium (Qs15). Qs15 showed the exponential phase up to 48hrs but in the presence of xylene it showed log phase up to 72hrs (Figure 4). Availability of nutrients is rate-limiting factor for successful bioremediation of hydrocarbons contaminated environments. Along with easily metabolized carbon source, microorganisms require various minerals like nitrogen, potassium, and iron for normal metabolism and growth. During the biodegradation of hydrocarbons as time increases, chemical changes in hydrocarbon, production of bio-products such as organic acids and depletion of minerals from the medium occurred, which indirectly affect the growth of bacteria [14].
Identification of the members of selected consortium
The BLAST search results depicted that the isolates Gh1 and Gh15 had the closest 16S rDNA sequence homology (99%) to Bacillus weihenstephanensis and Bacillus pumilus SAPR 032 respectively (Figure 5). The 16S rRNA gene partial sequence of strain Gh1 and Gh15 were submitted to the NCBI GenBank database under accession no KM385425 and KM385426 respectively. Thus, the constituents of the selected consortium were Bacillus sp. E14 and Bacillus sp. E87. ). The genus Bacillus is outstanding bacterial genus reported as petroleum hydrocarbon degrader. Their ability to form spores when nutrients are limiting makes species of Bacillus self-sustainable bioremediation tools [15].
Figure 5:Dendogram of Gh1 and Gh15 using PAST software showing 99% homology with Bacillus weihenstephanensis and Bacillus pumilus SAPR 032 respectively.

Biodegradation efficiency of Qs15
Figure 6:Degradation of selected hydrocarbon by Qs15 as assessed by GC-FID.
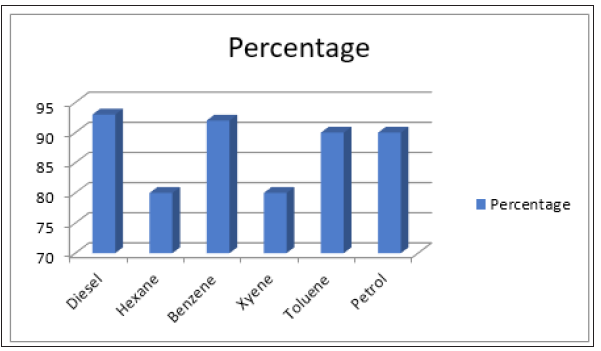
According to the GC-FID results, Qs15 exhibits the high biodegradation efficiency towards the hydrocarbons (Figure 6). QS15 showed the high degradation capability in aromatic hydrocarbons as compare to alkane in the medium. In presence of petrol and toluene in media, Qs15 degrades these two hydrocarbons up to 90%. Similar finding was revealed in the study of Sumathy [16] in which Pseudomonas spp. degrade the 1% toluene and result of GC-MS confirm the presence of catechol. Presence of catechol indicates the m-cleavage pathway of toluene degradation. Bacillus methylotrophicus contain plasmid-encoded operons account for the entire metabolic process that leads to biodegradation of xylene. Enzymes encoded by upper operon convert m-xylene into 3-methylbenzoate or benzoate respectively, and the enzyme of lower operon convert 3MBz into 3-methylcatechol or benzoate into catechol and it is the resultant of meta ring cleavage, which ultimately leads to intermediates of the TCA cycle [13]. On the basis of GC profile, it was depicted that Qs15 degrades 93% diesel within 48hrs as compared to control. Lima et al. [17] concluded that Bacillus sp. show the low degradation ability towards alkane present in diesel compared to higher alkane of diesel because short-chain n-alkanes dissolve the cellular membrane and are toxic to bacteria and long-chain n-alkanes have low solubility, inhibiting degradation by bacteria. In the degradation of benzene Qs15 achieved the 94% biodegradation efficiency while in case of hexane Qs15 showed 80% degradation that may be due to the induction of hexane-degrading enzymes that had already been induced during the preculture of Qs15 in the hexane-containing minimal salt medium. The enzymes alkane 1-monooxygenase, alcohol dehydrogenase, cyclohexanol-dehydrogenase, methane monooxygenase and cyclohexanone 1,2 monooxygenase are involved in degradation of alkanes [18]. Lee et al. (2010) found the level of 14C was higher in the medium supplemented with hexane (liquid phase) as compared to control, possibly due to the conversion of hexane into soluble metabolites by bacterium, EH831.
Individual bacteria could metabolize only to a limited hydrocarbons and the complete biodegradation requires mixture of different bacterial groups. The broad mineralization capacity exhibited by the mixed culture in this study is not surprising and may be attributed either to the presence of different microbial species with a number of metabolic pathways and/or to interspecies interactions [19]. Mixed populations have broad enzymatic capacities, which are responsible for the degradation of hydrocarbon at greater rate [20]. Microbial communities present in natural ecosystems proliferate together in a synergetic relationship and produce a remarkable cocktail of primary and secondary bioactive molecules (metabolites) including oxidative and hydrolytic enzymes, which are direct or indirect support the degradation of hydrocarbon [21].
Conclusion
The results of this study indicated that hydrocarbon-degrading bacteria isolated from oil contaminated soil, oil refinery, Assam, India, two potential strain (Bacillus sp. E14 and Bacillus sp. E87) were selected for the consortium construction (Qs15). have the highest potential to use different hydrocarbons (diesel, hexadecane, benzene, toluene and xylene) as the sole carbon source. This consortium Qs15 has high levels of crude oil degradation and sufficient growth on mineral medium supplemented with hydrocarbons. From the data of present investigation, it can be concluded Qs15 could be considered as good prospects for their application in bioremediation and restoration of hydrocarbon contaminated soil [22,23].
Acknowledgement
This work was supported by Maulana Azad National Fellowship, UGC, Delhi, India.
References
- Farber R, Rosenberg A, Rozenfeld S, Banet G, Cahan R (2019) Bioremediation of artificial diesel-contaminated soil using bacterial consortium immobilized to plasma-pretreated wood waste. Microorganisms 7(11): 497.
- Dombrowski N, Donaho J, Gutierrez T (2016) Reconstructing metabolic pathways of hydrocarbon-degrading bacteria from the deepwater horizon oil spill. Nature Microbiology 1(7): 16057.
- Gurav R, Lyu H, Ma J, Tang J, Liu Q, et al. (2017) Degradation of n-alkanes and PAHs from the heavy crude oil using salt-tolerant bacterial consortia and analysis of their catabolic genes. Environmental Science Pollution Res 24(12): 11392-11403.
- Xiao Y, Chen S, Gao Y, Hu W, Hu M, et al. (2015) Isolation of a novel beta-cypermethrin degrading strain Bacillus Subtilis BSF01 and its biodegradation pathway. Applied Microbiology and Biotechnology 99(6): 2849-2859.
- Boboye B, Olukunle OF, Adetuyi FC (2010) Degradative activity of bacteria isolated from hydrocarbon-polluted site in Ilaje, Ondo State, Nigeria. African Journal of Microbiology 4(23): 2484-2491.
- Kumar G, Singla R, Kumar R (2010) Plasmid associated Anthracene degradation by Pseudomonas sp. isolated from filling station site. Nature and Science 8(4): 89.
- Trevors JT (1986) Plasmid curing in bacteria. FEMS Microbiology Review 32(3-4): 149-157.
- Lowry OH, Rosenbrough NH, Farr AL, Randall RJ (1951) Protein measurement with the folin-phenol reagent. Journal of Biological Chemistry 193(1): 265-275.
- Bazzicalupo M, Fani R (1995) Methods in molecular biology, species diagnostic protocols: PCR and other nucleic acid methods. In: Clap JP (Ed.), Humana Press Inc., Totowa NJ, USA. pp.112-124.
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology & Evolution 24(8): 1596-1599.
- Naggar AY, Kamel MM, Hassan YM, Youssel Kh A, Al Adly AA, et al. (2012) Gas chromatographic assessment for bioremediation of hydrocarbons pollutants using Bacillus Amyloliquefaciens. Archive of Applied Science Research 4(4): 1593-1599.
- Borah D, Yadav RNS (2015) Plasmid curing of a novel hydrocarbon degrading Bacillus cereus Strain DRDU1 revealed its involvement in petroleum oil degradation. Journal of Petroleum & Environmental Biotechnology 6(3): 1-4.
- Undugoda LJS, Kandisa RV, Kannangara S, Sirisena DM (2018) Plasmid encoded toluene and xylene degradation by phyllosphere bacteria. Journal of Environmental & Analytical Toxicology 8(2): 1-4.
- Ghosal D, Ghosh S, Dutta TK, Ahn Y (2016) Current state of knowledge in microbial degradation of polycyclic aromatic hydrocarbons (PAHs): A review. Frontiers in Microbiology 7: 1369.
- Oyetibo GO, Chien MF, Ikeda Ohtsubo W, Suzuki H, Obayori OS, et al. (2017) Biodegradation of crude oil and phenanthrene by heavy metal resistant Bacillus subtilis isolated from a multi-polluted industrial wastewater creek. International Biodeterioration & Biodegradation 120: 143-151.
- Sumathy VDH (2017) Isolation of toluene degrading bacteria from polluted environment. European Journal of Pharmaceutical and Medical Research 4(9): 341-346.
- Lima SD, Oliveira AF, Golin R, Lopes VCP, Caixeta DS, et al. (2018) Isolation and characterization of hydrocarbon-degrading bacteria from gas station leaking-contaminated groundwater in the Southern Amazon, Brazil. Brazilian Journal of Biology 80(2): 354-361.
- Bacosa HP, Erdner DL, Rosenheim BE, Shetty P, Seitz KW, et al. (2018) Hydrocarbon degradation and response of seafloor sediment bacterial community in the northern Gulf of Mexico to light louisiana sweet crude oil. Journal of the International Society for Microbial Ecology 12(10): 2532-2543.
- Handayani S, Safitri R, Surono W, Astika H, Damayanti R, et al. (2019) Biodegradation of BTEX by indigenous microorganisms isolated from UCG project area, South Sumatra. IOP Conf Ser: Earth Environ Sci 308: 1-11.
- Jarallah EM, AL Ritha FTA, Jabuk SIA, Abdullah ZA (2014) Detection and evaluation of hydrocarbon mixtures degrading bacteria isolated from desiel polluted soils. Journal of Babylon University/Pure and Applied Sciences 4(22): 1-12.
- Truskewycz A, Gundry TD, Khudur LS, Kolobaric A, Taha M, et al. (2019) Petroleum hydrocarbon contamination in terrestrial ecosystems-fate and microbial responses. Molecules 24(18): 3400.
- Al Wasify RS, Hamed SR (2014) Bacterial biodegradation of crude oil using local isolates. International Journal of Bacteriology
- VanDecar JC, Russo RM, James DE, Ambeh WB, Franke M (2003) Aseismic continuation of the lesser antilles slab beneath northeastern Venezuela. Journal of Geophysical Research 108(1): 1-15.
© 2020 Eram Ayaz. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






