- Submissions

Full Text
Significances of Bioengineering & Biosciences
Study of the Binding Site of Long Chain Fatty Acids on Fatty Acid-Binding Proteins.
Tarhda Z*
Biotechnology lab (MedBiotech), Morocco
*Corresponding author:Zineb Tarhda, Biotechnology lab (MedBiotech), Faculté de Médecine et de Pharmacie de Rabat, Morocco
Submission: August 08, 2018; Published: April 26, 2019

ISSN 2637-8078Volume3 Issue1
Abstract
Fatty acid binding proteins (FABPs) are proposed to function in the transport, metabolism, or storage of free fatty acids. Despite extensive studies, the mechanism of fatty acid binding and the ability of FABPs to discriminate among fatty acids according to their chain lengths and saturation state are poorly understood. In this work, we aimed to study the key features of FABP binding site based on the crystallographic structure of FABP-LCFA complexes, sequence multiple alignment and structural superposition of some FABPs. Due to the important role of the physicochemical features of the proteins in the ligand interaction, we studied those of the FABPs. The nature of the bond between the FABPs and the different LCFA, also the part of LCFA involved in LCFA-FABP interaction is interesting to detect the mechanism of LCFAs binding on FABPs. To achieve these objectives, we used different databases and various bioinformatics programs. Based on the results, we were able to answered some unclear about the mechanism of fatty acid binding.
Keywords: Long chain fatty acids; Fatty acid binding site; Multiple alignment; Similarity; Physicochemical characteristics
Introduction
FABPs carry FAs through the aqueous cellular environment and are involved in processes like FA uptake, transport and oxidation. Their genes consist of 4 exons and 3 introns [1], and predominantly expressed in different tissues and the protein are named accordingly:
a) Liver-FABP (L-FABP)
b) Intestine-FABP (I-FABP)
c) Heart-FABP (H-FABP)
d) Adipocyte-FABP (A-FABP)
e) Epidermal-FABP (E-FABP)
f) Ileal-FABP (Il-FABP)
g) Brain-FABP (B-FABP)
h) Myelin-FABP (M-FABP)
i) Testis-FABP (T-FABP)
FABPs demonstrate strong evolutionary conservation [2,3] and are present in a spectrum of species including Drosophila melanogaster, Caenorhabditis elegans, mouse and human [3]. Each individual FABP has its own sequence, but all share a common structure consisting of ten antiparallel β-barrel (βA-βJ) structures containing a solvent-accessible ligand binding pocket, capped by an N-terminal helix-turn-helix motif (αI-αII) which is thought to act as the regulatory portal for binding [4-7]. All FABPs are capable of binding long-chain fatty acids (LCFAs, C12-20), which differ in their selectivity, affinity and binding mechanism [4,8]. In addition to the reported works on the FABPs, we performed this study designed to assemble the maximum information about the FABPs and especially to extract new knowledge about the characteristics of the LCFA-FABP interaction mechanisms. The tertiary structure of LCFA bound to FABP was determined by X-ray diffraction and/or NMR, among these FABP we find FABP-H, FABP4-Mouse, FABP-B, FABP-SCHMA, FABP-LOCIMI, FABPAMBE, FABPL-RAT, FABP-L. Every one of these FABP is able to bind more than one molecule of LCFA in the same time and saturated or unsaturated fatty acid state.
In this study, we chose five long chain fatty acids and we classified a number of FABPs according to these LCFAs. Besides, we determined the binding site for each LCFA included in this study and made a head to head comparison between the active sites of the same FABP. The nature of the bond between the FABPs and the LCFAs, the binding site residues of FABPs and the part of LCFA included in this bond are useful for understanding the mechanism of FABP-LCFA interaction. Multiple alignments and structural superposition were used to demonstrate the consensus residues of the FABPs and the residues of the FABPs playing a crucial role in the LCFA binding. As it was expected, the interaction of the protein with the ligands depends on ligand’s and protein’s physicochemical characteristics.
Methods
Fatty acid binding proteins categories
We used Uniprot database and Protein Data Bank in Europe Bringing Structure to Biology [9,10] that provides the information on protein and fixed ligand.
Fatty acid binding site residues
Active site residues of the FABPs already mentioned above are extracted from PDBeMotif database [11] and Jena Library of Biological Macromolecules [12]. The determination of the active site allows the study of the nature of residue-FA bond and the LCFA part involved in FABP interaction.
Comparison between the active sites of the same FABP
The comparison of the active site components of the same FABP is interesting to determine if there are any FABPs key residues for the FA binding and to deduce the origin of the FABPs specificity for each FA.
Multiple alignments and structural superposition of FABPs
The ClustalW2 multiple sequences alignment software program [13] is used to perform the multiple alignments of each FABP category. The structural superposition of FABPs in the same class is done by Pymol tool which enabled to visualize the differences between the FABPs binding sites.
Physicochemical characteristics of FABPs
Biological activity of proteins depends on their physicochemical characteristics, hence the importance to study those of FABPs. Extinction coefficients, Instability index (II), Aliphatic index, GRAVY (Grand Average of Hydropathy), Molecular weight and theoretical pI (Isoelectric point) are the physicochemical characters of FABPs were computed using the Expas’s prot-param tool [14]. In addition, we determined the number and position of the cysteine residue to study disulfide bonds in FABPs using CYS-REC server [15].
Results
Fatty acid binding sites study
According to the databases used, we chose the following FABPs represented by their PDB codes:
a) FABPH: 1HMS, 1HMR, 1HMT
b) FABP4_MOUSE: 1LIE, 1LIF, 2Q9S, 1ADL
c) FABP7_HUMAN: 1FE3
d) FABP_SCHMA: 1VYF, 1VYG
e) FABPM_LOCMI: 2FLJ
f) FABPI_RAT: 2IFB
g) FABPL_RAT: 2JU8
h) FABPL_HUMAN: 2LKK, 3STM
i) FABP5-HUMAN: 1B56
j) FABP1_ECHGR: 1O8V
These FABPs bind oleic acid, stearic acid, arachidonic acid, palmitic acid and linoleic acid are subjected to study the binding site of LCFAs and the mechanism of ligand-receptor interactions. The FABPs bind either one or two ligands in a single binding site as it shall bind different FAs separately. In this study, only the FABPs binding a single FA molecule in their binding sites.There are five FABP categories depending on the bound LCFA:
a) Oleic acid binding proteins ‘OABPs’: (PDB codes) 1FE3, 1HMR (Figure 1), 1HMS, 2FLJ, 2ju8, 1VYF, 2LKK. (2) Arachidonic acid binding proteins ‘AABPs’: (PDB codes) 1VYG, 1ADL.
b) Stearic acid binding proteins ‘SABPs’: (PDB codes) 1HMT, 1LIF.
c) Linoleic acid binding protein ‘LABPs’: (PDB codes) 2Q9S.
d) Palmitic acid binding proteins ‘PABPs’: (PDB codes) 1O8V, 1B56, 1LIE, 2IFB, 2HMB, 3STM.
Figure 1:X-Ray illustration of FABP-Oleic acid binding site (PDB code: 1HMR) visualized by Pymol..
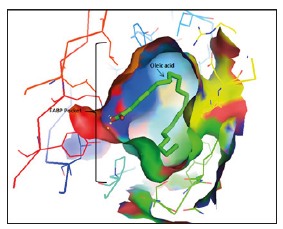
The active sites of FABP are grouped below:
a) Visualization of oleic acid binding site
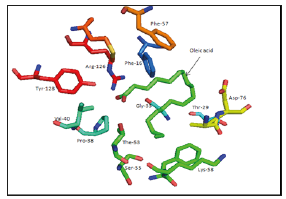
I. 1FE3: Phe-16, Thr-29, Gly-33, Pro-38, Val-40, Thr-53, Ser- 55, Phe-57, Lys-58, Asp-76, Arg-126, Tyr-128 (Figure 2).
II. 1HMR: Ala-33, Thr-53, Lys-58, Thr-60, Asp-76, Leu-115, Arg-126, Tyr-128.
III. 1HMS: Met-20, Ala-33, Lys-58, Leu-115, Arg-126, Tyr-128 [16,17].
IV. 2FLJ: Phe-17, Tyr-20, Met-21, Ile-24, Val-26, Ala-33, Ile- 59, Leu-77, Arg-108, Ile-117, Arg-128, Tyr-130.
V. 1VYF: Met-20, Gly-33, Thr-53, Ser-55, Phe-57, Lys-58, Ser- 75, Asp-76, Arg-107, Arg-127, Tyr-129.
VI. 2JU8: Ile-41, Phe-63, Leu-71, Val-83, Thr-102, Ile-109, Arg-122.
VII. 2LKK: Ser-39, Ile-52, Leu-71, Phe-95, Thr-102, Asn-111, Met-113, Arg-122, Ser-124.
Figure 2:Oleic acid binding site of 1FE3 as an example of the binding..
b) Visualization of arachidonic acid binding site
I. 1VYG: Phe-16, Met-20, Leu-23, Val-25, Gly-33, Pro-38, Ser- 55, Lys-58, Ser-75, Asp-76, Arg-78, Ile-105, Arg-107, Thr-116, Arg- 127, Tyr-129.
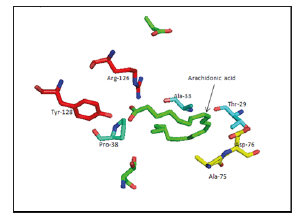
II. 1ADL: Thr-29, Ala-33, Pro-38, Ala-75, Asp-76, Arg-126, Tyr-128 (Figure 3).
Figure 3:Arachidonic acid binding site of 1ADL as an example of the binding.
c) Visualization of stearic acid binding site
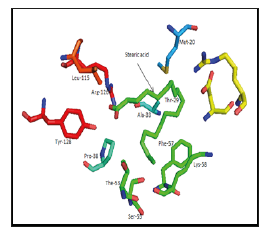
I. 1HMT: Met-20, Ala-33, Thr-53, Ser-55, Phe-57, Lys-58, Leu-115, Arg-126, Tyr-128 (Figure 4); [18].
Figure 4:Stearic acid binding site of 1HMT.
II. 1LIF: Met-20, Val-32, Ala-33, Met-40, Phe-57, Lys-58, Ala- 75, Asp-76, Cys-117, Arg-126, and Tyr-128.
d) Visualization of linoleic acid binding site
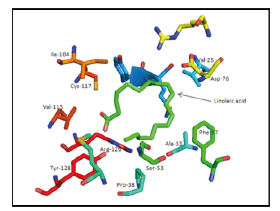
I. 2Q9S: Val-25, Ala-33, Pro-38, Ser-53, Phe-57, Asp-76, Ile- 104, Val-115, Cys-117, Arg-126, Tyr-128 (Figure 5).
Figure 5:Linoleic acid binding site of 2Q9S as an example of the binding.
e) Visualization of palmitic acid binding site
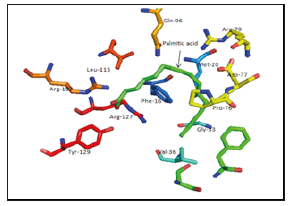
I. 1O8V: Phe-16, Met-20, Gly-33, Val-36, Pro-76, Asp-77, Arg-79, Gln-96, Arg-107, Arg-127, Tyr-129 (Figure 6).
Figure 6:Palmitic acid binding site of 1O8V as an example of the binding.
II. 1B56: Met-35, Gly-36, Asp-79, Arg-109, Val-118, Arg-129, Tyr-131.
III. 1LIE: Thr-29, Ala-33, Met-40, Lys-58, Ala-75, Asp-76, OCS 117, Arg-126, Tyr-128.
IV. 2IFB: Tyr-14, Val-49, Phe-62, Trp-82, Arg-106 [19].
V. 2HMB: Ala-33, Arg-126, Tyr-128.
VI. 3STM: Ser-39, Phe-50, Glu-72, Thr-102, Asn-111, Arg-122.
The analysis of the FABP residue-FA bond nature shows in Table 1. The bonds type between FABPs and palmitic acid are not present in the PBDEs Motif database. The software tool Poseview helped to determine some interactions between PPFAs and palmitic acid (Figure 7). The analysis of LCFA parts involved in the FABP interaction showed that all LCFAs are bound with their ligand by specific parts: Carboxyl group, C8-C10, C12-C13, C13-C16, C15-C18.
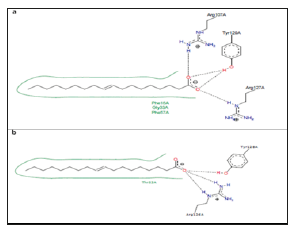
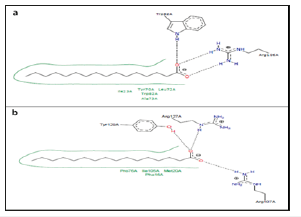
Figure 7:Two-dimensional diagrams of OABPs- Oleic acid complexes. (a) Complex of FABP_SCHMA with oleic acid. PDB code 1VYF. (b) Complex of FABPH with oleic acid. PDB code 1HMR.
f) Oleate: 18 C, 1 double bond C9-C10
Table 1:The interaction types of some FABPs with the FA.
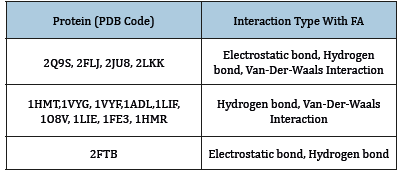
Figure 7 shows the OABPs-Oleic acid complexes.
Carboxyl group: The last Tyr and Arg that proceeds.
I. C1: Leu-115, Thr-116, Ile-117.
II. O1: Arg-107 and 108.
III. C9-C10: Met-73 or Asp-76 or [leu-77+Met-20] or Met-20 (Val is frequent at this level).
IV. C12-C16: Gly-33 or [Ala-33+Ser-73] or Ser-55, Phe-57 and Phe-95 also are frequent.
V. C17-C18: Lys-58 or [Lys-58+Thr-53] or [Lys-58+Phe-57] or Ile-59.
g) Arachidonate: 20C, 4 double bonds (C6-C7, C9-C10, C12-C13, C15-C16)
Carboxyl group: The last Tyr and Arg that precedes, Arg107.
I. C9-C11: Asp-76.
II. C12-C16: Gly-33 and [Ala-33+ Met-20].
III. C18: Pro-38.
IV. C18-C20: [Ser-55 +Lys-58] at level of C20.
h) Stearate: 18C
Carboxyl group: The last Tyr and Arg that precedes.
I. C8-C10: Asp-76 or [Met-20 +Asp-76].
II. C12-C13: Ala-33.
III. C13-C16: Phe-57.
IV. C15-C18: Lys-58 or [Lys-58+Ser-55].
i) Linoleic acid: 18 C, 2 double bonds (C6-C7, C9-C10)
Carboxyl group: [Tyr-128+Cys-117] and Val-115.
I. C5: Ile-104.
II. C9-C10: Asp-76, Val-76
III. C12-C13: Ala-33 and Phe-57.
IV. C15-C16: Pro-38 and Arg-126.
V. C17-C18: Ser-53.
An exemple of LABP-Linoleic acid interaction is showed in Figure 8-10.
j) Palmitic acid: 16 C
Figure 8:Two-dimensional diagrams of AABPs- Arachidonic acid complexes. (a) Complex of FABP_SCHMA with arachidonic acid. PDB code 1VYG. (b) Complex of FABP4_MOUSE with arachidonic acid. PDB code 1ADL.
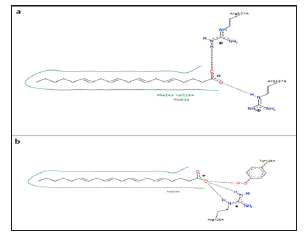
Figure 9:Two-dimensional diagrams of SABPs- Stearic acid complexes. Example of FABP4_ mouse-stearic acid complex. PDB code 1LIF.
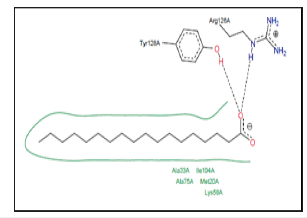
Figure 10:Two-dimensional diagrams of LABPs_ Linoleic acid complexes. Example of FABP4_ MOUSE-linoleic acid complex. PDB code 2Q9S.
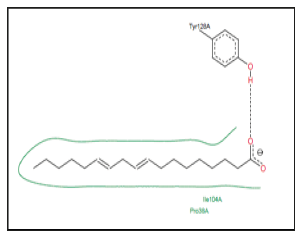
The distribution of binding site residues around palmitic acid are missed in PDBemotif database but ViewPower provided an idea about the basic residues which form the binding site and some interactions. Figure 11 shows the prediction interaction between palmitic acid and some PABP. The comparison between the active sites of the same FABP showed in Table 2. The active site of FABPSCHMA characterized by the intervention of a relatively large number of residues compared to the other FABPs. FABPL-Rat has a very high similarity to FABPL-Human, it is involved in the binding of oleic acid by residues, which common with those of 2LKK are fixing the same ligand. Saturation state, double bond position and the length of FA has no effect on the mechanism of their attachment to FABP which is clear when we compared between the active site of the same FABP for binding of more than one type of FA this complicates the ability of FABPs to discriminate FAs.
Table 2:Principal residues of FABPs binding sites.
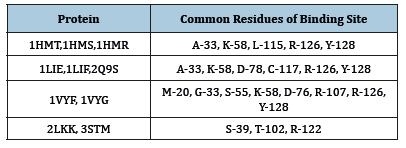
Figure 11:Two-dimensional diagrams of PABPs- Palmitic acid complexes. (a) Complex of FABPI_RAT with palmitic acid. PDB code 2IFB. (b) Complex of FABP1_ECHGR with palmitic acid. PDB code 108V.
Multiple alignment and structural superposition of the FABP categories
Figure 12:Multiple alignment of oleic acid binding proteins.
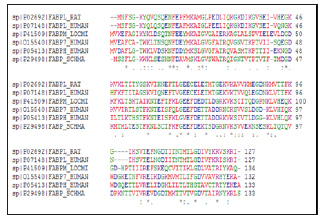
Multiple alignment and structural superposition of OABPs Figure 12 showed: In the active site of OABP, there are necessary residues which are retained like the case of Phe-63, and there are other not conserved in the OABPs, such as Met-20 and Lys-58 which keep the same position at all OABP.
a) Asp-76 is conserved in OABP except at 2LKK, 1VYF.
b) Phe-57 occupies the same position in 1HMS, 1VYF and 1FE3, these proteins contain a conserved Ser-55 dominant also in 2LKK and 2JU8.
c) Gly-33 and Ala33 are semi-conservative in the OABPs where they play a significant role in their active sites. Leu-115, Thr- 116 and Ile-117 are involved in the binding of OA in 1HMS, 1VYF and 2FLJ respectively, these residues occupy the same position according to the multiple alignment.
d) Leu-77, Met-73 and Ser-75 occupy the same position in 2FLJ and 1VYF.
e) Arg-107 keeps the same position in 1FE3, 1HMS, 1VYF and 2FLJ.
f) Arg-126 is conserved in the OABPs; it is numbered 122 in 2JU8, 2LKK.
g) Tyr-128 of 2JU8 and 2LKK keeps the same position in the OABPs.
h) Gly-33/ Ala-33, Asp in position 76, the last Tyr and Arg that preceding directly are the most important residues in the active site of AOBP. In addition we have detected that other residues play a crucial role, we should consider them as additional residues: Ser- 55, Lys-58, Met-20, Met-73, Ser-72, and Arg-107. FABPL-Human and FABPL- Rat binding to AO by the last Arg, Leu-71 and Thr-102, these residues are essential for this uptake (Figure 12).
Arg 127, Tyr 129, Arg107, Asp 76, Pro38, Ser 55, Lys58 have a significant role in AA uptake. Except of Gly33 and Ala33 which are semi-conservative, all the amino acids mentioned above are preserved in AABPs (Figure 13). The residues constituting the active site mentioned above are conserved in both SABPs (Figure14). FABPH-Human and FABP4-Mouse have the higher similarity by 64%. Gly 33, Met20, Arg 107, Asp 77, the last Tyr and Arg preceding preserved in the majority of PABPs and represent the active site of palmitic acid binding, Ala33 and Gly33 are semi-conserved in PA binding (Figure15). It is deduced that Ala/Gly in 33 position, Asp occupies 76 position, the last Tyr and Arg that proceeds of FABPs are essential in binding of LCFAs. Lys58, Ser55, Met20, Met73, Pro38, Cys117 and Val play a complementary role in the active site of FABPs.
Figure 13:Structural superposition of 1VYG and 1ADL.
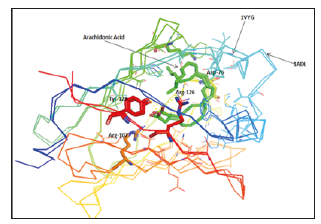
Figure 14:Multiple alignment of stearic acid binding proteins.
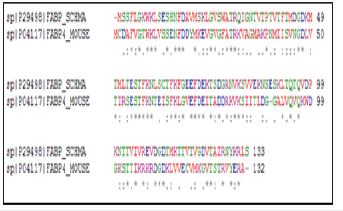
Figure 15:Multiple alignment of palmitic acid binding proteins.
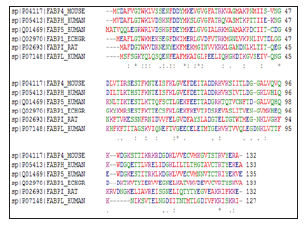
Determination of physicochemical characteristics of FABPs
The computed pI value for FABPH-Human, FABP7_Human, FABPM_LOCMI, FABP5_Human, FABPI_Rat, FABPL_Human (pI<7) indicated their acidic nature, whereas pI for FABP_SCHMA, FABP4_ Mouse, FABP1_ECHGR (pI>7) revealed there basic behavior. On the basis of instability index Expasy’s ProtParam classified the FABPs as stable (Instability index<40). The aliphatic index (AI) which is defined as the relative volume of a protein occupied by aliphatic side chain is regarded as the positive factor for the increase of thermal stability of globular protein. The high aliphatic index of all FABPs infers that these proteins may be stable for a wide range of temperature. The very low GRAVY index of FABPs infers that these proteins could result in a better interaction with water (Table 3). As percentage of Cysteine residue is very low in all the FABPs under study, none of these proteins have disulphide bond linkages, as indicated by CYS-REC result (Table 4).
Table 3:Presence of disulphide (SS) bond of FABPs predicted by Cys-Rec serve
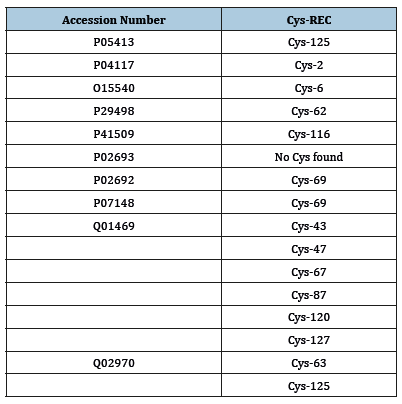
Table 4:Physicochemical characters of FABPs as predicted by Expas’s prot-param program.

Discussion
Quantitative analysis of the interaction of the ligand with the FABP structure clearly implicates both portal and anti-portal domains as the preferred domains [20], which we have concluded in the present study. It is likely that a conformational change in the portal region occurs during FA binding or release [21]. Fatty acid binding mechanism still remains mysterious and each study has proposed an interpretation based on their results. According to Banaszak, the cavity differs in molecular detail which is likely to be an important factor for the observed differences in the affinity and specificity of different FABPs. Other studies also suggest since the cavity surface is very rough, one might expect that small conformational changes of the side chains lining the cavity could result in a substantive increase in the cavity hydrophobic profile. In fact, such changes may be an important part of the ligand binding mechanism [22].
In this study we have deduced the cavity of FABPs is irrelevant in the affinity and the specificity for the FA. The superposition of some FABP fixing different ligands shows an uniformity of their conformations and their cavities, the example of [1VYG, 1VYF]; [1HMS, 1HMT] and [1ADL, 1LIE, 1LIF]. However, we have noticed a difference in the cavities of some FABP from the same family, the case of 2Q9S comparing with 1ADL, 1LIE and 1LIF. 2Q9S is characterized by a relatively small cavity, due to the Phe-56 oriented toward the cavity inside reducing its volume. The same for 3STM and 2LKK in which the Phe-95 affects the cavity, it is long in 2LKK and reduced in 3STM. Also, the superposition of different FABP fixing the same FA shows a huge difference in their cavities. The ligand affinity can be correlated with the conformational energy and the solubility of the bound ligand. This finding could be related to the ability of FABPs to discriminate based on fatty acid chain length and saturation state [23].
In our study we have interested by the analysis of the FA affinity to FABPs according to the saturation state and/or the chain length. We have concluded the two criteria have no effect on the FABP binding because some FABPs studied could bind a saturated FA with short chain as they should fix an unsaturated FA with long chain. For all crystalline structures, the ligands are fitted snugly inside the inner cavity of the protein with some variation in their orientation depending on the type of the protein (liver, intestinal, etc.) [20]. According to our study based on the databases, and the results of experimental researches done in this context, in the FABPs studied, the last Tyr and Arg which above are directly involved in the LCFAs binding (O1 and/or O2) by hydrogen bond. It confirms as noted previously, all FABPs bind FA with its carboxylate group oriented inside the cavity interacting with interior Arg residues and other side chain [24]. Based on our analysis, a specific part of LCFA involved in the FABP-FA interaction, also some residues when acting they interact with the FA on the same level, there is the last Tyr and Arg that precedes interacting with carboxyl group, Ala-33 or Gly-33 which still interact with the C12-C16 part, Asp-76 with the field C8-C10, and Lys-58 with the last part of the FA.
It is deduced the FABPs act with the FA indifferently. The similarity between FABPs has no relationship to the fixed FA. The exceptions of pH, all of the physicochemical parameters of FABPs studied are widely close. The water molecule is routinely present inside the protein, it found in the FABP-binding cavity and intrinsically involved in the binding process [25,26]. The water considers essential in the fixing of LCFA inter the cavity to which it bind by Vander-Der-Wals force, hydrogen bond o electrostatic bond. Double bond in FA, its position and the chain length of FA has no effect on the attachment mechanism to FABP. So the interaction between the FABP and the different FA can be controlled by the pH of FABP, their environment and/or their tissue distribution [27].
Conclusion
To identify the key features of FA-FABP interaction, we have performed an exhaustive analysis of the FABPs in which we have recruited a number of FABPs data, we have analyzed the structure crystallized of FABP complexes, realized the sequence multiple alignment and the structural superposition and we have determined physicochemical parameters. Successfully, we have validated some hypotheses based on experimental studies and we have corrected else. This study provides a new compared to reported work such as the determination of FABP essential residues implicate in the FA binding and the factors which play a crucial role in this binding.
References
- Sweetser DA, Lowe JB, Gordon JI (1986) The nucleotide sequence of the rat liver fatty acid-binding protein gene. Evidence that exon 1 encodes an oligopeptide domain shared by a family of proteins which bind hydrophobic ligands. J Biol Chem 261(12): 5554-5561.
- Patrick BJ (2009) Molecular evolution of vertebrate fatty-acid binding proteins. pp. 17-29.
- Smathers RL, Petersen DR (2011) The human fatty acid-binding protein family: Evolutionary divergences and functions. Hum Genomics 5(3): 170-191.
- Chmurzynska A (2006) The multigene family of fatty acid-binding proteins (FABPs): Function, structure and polymorphism. J Appl Genet 47(1): 39-48.
- Liou HL, Kahn PC, Storch J (2002) Role of the helical domain in fatty acid transfer from adipocyte and heart fatty acid-binding proteins to membranes: analysis of chimeric proteins. J Biol Chem 277(3): 1806-1815.
- Lucke C, Rademacher M, Aukje WZ, Van Moerkerk JH, Rüterjans H (2001) Spin-system heterogeneities indicate a selected-fit mechanism in fatty acid binding to heart-type fatty acid-binding protein (H-FABP). Biochem J 354(Pt 2): 259-266.
- Zanotti SG, Scapinj G, Spadon P, Veerkampq JH, SacchettiniQ JC (1992) Three-dimensional structure of recombinant human muscle fatty acid- binding protein. J Biol Chem 267(26): 18541-18550.
- Furuhashi M, Hotamisligil GS (2008) Fatty acid-binding proteins: Role in metabolic diseases and potential as drug targets. Nat Rev Drug Discov 7(6): 489-503.
- http://www.uniprot.org/
- http://ebi.ac.uk/pdbe-srv/view/
- http://ebi.ac.uk/pdbe-site/pdbemotif/
- http://imb-jena.de/IMAGE.html
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21): 2947- 2948.
- http://web.expasy.org/protparam/
- http://linux1.softberry.com/berry.phtml?topic=cys_rec&group=programs& subgroup=propt
- Woolf TB, Tychko M (1998) Simulations of fatty acid-binding proteins II sites for discrimination of monounsaturated ligands. Biophysical Journal 74(2): 694-707.
- Clemens FMP, Veerkamp JH (1996) Fatty acid binding and conformational stability of mutants of human muscle fatty acid-binding protein. Biochem J 314( Pt 1): 253-260.
- Woolf TB (1998) Simulations of fatty acid-binding proteins suggest sites important for function I stearic acid. Biophys J 74(2 Pt 1): 681-693.
- Mihajlovic M, Lazaridis T (2007) Modeling fatty acid delivery from intestinal fatty acid binding protein to a membrane. Protein Sci 16(9): 2042- 2055.
- Levin LB, Ganoth A, Amram S, Nachliel E, Gutman M, et al. (2010) Insight into the interaction sites between fatty acid binding proteins and their ligands. J Mol Model 16(5): 929-938.
- Storch J, McDermott L (2009) Structural and functional analysis of fatty acid-binding proteins. J Lipid Res S126-S131.
- Likić VA, Prendergast FG (1999) Structure and dynamics of the fatty acid binding cavity in apo rat intestinal fatty acid binding protein. Protein Sci 8(8): 1649-1657.
- Young AC, Scapin G, Kromminga A, Patel SB, Veerkamp JH, et al. (1994) Structural studies on human muscle fatty acid binding protein at 1.4 Å resolution: Binding interactions with three C18 fatty acids. Structure 2(6): 523-534.
- Richieri GV, Ogata RT, Kleinfeld AM (1999) Fatty acid interactions with native and mutant fatty acid binding proteins. Mol Cell Biochem 192(1- 2): 77-85.
- Likić VA, Juranić N, Macura S, Prendergast FG (2000) A structural water molecule in the family of fatty acid binding proteins. Protein Sci 9(3): 497-504.
- Tsfadia Y, Friedman R, Kadmon J, Selzer A, Nachliel E, et al. (2007) Molecular dynamics simulations of palmitate entry into the hydrophobic pocket of the fatty acid binding protein. FEBS Letters 581(6): 1243-1247.
- http://poseview.zbh.uni-hamburg.de/
© 2019 Tarhda Z. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






