- Submissions

Full Text
Significances of Bioengineering & Biosciences
Genome-Centric Evaluation of Bacillus sp. strain –ATCC55673 and Response to Uranium Biomineralization
Victor Ibeanusi1*, Ashish Pathak1*, Ashvini Chauhan1, Jada Hoyle-Gardner1, Tyrik Cooper1, Landon Turker1, Harley Howard1, Oluchukwu Obinegbo1, Gang Chen2 and John Seaman3
1 Florida A&M University, USA
2 Department of Civil & Environmental Engineering, FAMU-FSU College of Engineering, USA
3 University of Georgia, Aiken, USA
*Corresponding author:Victor Ibeanusi, School of the Environment, 1515 S Martin Luther King Jr Blvd, Suite 305B, FSH Science Research Center, Florida A & M University, Tallahassee, 32307, Florida, USA
*Corresponding author:Ashish Pathak, School of the Environment, Florida Agricultural and Mechanical University, USA, Email:ashishpathak72@gmail.com
Submission: July 12, 2018; Published: September 12, 2018

ISSN 2637-8078 Volume2 Issue3
Abstract
A patented microbial systems process was conducted on microbial-mediated biodegradative mechanisms at a coal-pile run off basin discharged from the coal fired plant at Savannah River Site, Aiken, South Carolina (SC). One of the isolated strains, ATCC 55673, grew robustly on number of dissolved toxic metals. Genome-centric evaluation revealed the isolate to belong to the genus Bacillus with close affiliation to B. cereus, an aggressive polychlorinated biphenyl-degrader. At a coverage of 90x, the genome of strain ATCC 55673 consisted of 4,615,850 bases with a total number of 4,590 putative genes assembling into 51 contigs with an N50 contig length of 2,597,36 bases. Several gene homologues coding for resistance to heavy metals were identified, such as a suite of outer membrane efflux pump proteins like nickel/cobalt transporter regulators, peptide/nickel transport substrate and ATP-binding proteins, permease proteins, a high-affinity nickel-transport protein, and several genes for metabolism of aromatic compounds (Table 1).
Table 1:Specifications.

Keywords: Biomineralization of uranium and heavy metals, organic waste; Metal resistance genes; Whole genome sequencing (WGS); Bacillus sp. (ATCC 55673)
Introduction
Metals and radionuclides are released into the environment as a function of nuclear energy activities, accidental spills from nuclear power plants, the production of nuclear weapons and their testing (www.epa.gov/radiation/index.html). Inadvertent release of radionuclides represents a serious threat for both environmental and public health since they are easily integrated into the food chain through contaminated subsurface soil and groundwater. Because of historical contamination from nuclear materials manufacturing and processing, several sites managed by the US Department of Energy (DOE) remain co-contaminated by radionuclides, mainly Uranium (U). Such co-contaminated ecosystems are difficult to remediate using conventional approaches, such as excavation and disposal or pump-and-treat. Less expensive in-situ immobilization and remediation strategies rely on chemical or nutrient additives that alter contaminant speciation or enhance microbial activity, leading to enhanced solid-phase partitioning to reduce migration, bioavailability and ultimately the associated exposure hazard. In fact, the effectiveness of PO4 minerals and compounds in stabilizing many heavy metals and radionuclides through sorption and/or the formation of secondary phosphate precipitates has been successfully demonstrated. Despite the effectiveness of PO4 for metal and radionuclide decontamination, its application is limited due to: (1) the necessity of incorporating sparingly soluble, solid-phase PO4 sources within the contaminated sites, limiting application to shallow soils/sediments, (2) the long-term effectiveness of contaminant immobilization once PO4 addition has ceased, and (3) the impact of PO4 on co-contaminants such as 129I and 99Tc. To overcome solubility limitations, a range of soluble P materials including phytate and other soluble P sources have been evaluated with varied success [1-7].
This study focused on the Savannah River Site (SRS), which is an approximately 800-km2 former nuclear weapons production facility located along the Savannah River near Aiken, SC. SRS is of significant interest because of widespread contamination by residual heavy metals and radionuclides from the previous DOE weapons production activities. SRS environmental monitoring has revealed that the groundwater and riparian sediments contain uranium (U), nickel (Ni) and other metals or radionuclides, specifically in the M-Area. Tims Branch, which drains the upland region around the M area into Upper Three Runs and eventually the Savannah River, is a second-order stream that receives contamination from Steeds Pond, a former farm pond that served as a radiological setting basin. An estimated 43,500kg of U and a similar amount of Ni were released from the nuclear fabrication facility, known as M Area to the Tims Branch Steeds Pond system from 1966 to 1968 associated with target fabrications in M-Area, representing 97% of the gross alpha activity released by the SRS. Overall, the Beaver Ponds, surrounding wetlands, and former farm ponds have essentially been functioning as settling basins for heavy metals and radionuclides released into the stream system.
To advance microbial-mediated strategies for effective uranium biomineralization and heavy metals remediation, we report here the first genomic sequencing of ATCC 55673, a Bacillus sp. previously reported in Ibeanusi et al. [8] as a bacterial candidate for effective remediation of radionuclides, heavy metals, and volatile organic compounds.
Materials and Methods
Isolation of heavy metals and dissolved metals resistant strain ATCC 55673
Several bacterial strains resistant to high concentrations of heavy metals and various dissolved metals were isolated from activated sludge of the waste water treatment plant by serially diluting sludge slurries and plating onto Luria Bertani (LB) media supplemented with dissolved metals; these concentrations are like those present in the collected sludge [9-14]. Plates were incubated aerobically at 30 °C and colonies that grew within a week were further isolated on LB amended with Al, Ni, Zn, Cu, Fe and combinations thereof. ATCC 55673 was found to grow robustly on number of dissolved metals and some heavy metals and therefore was chosen for further genome-centric evaluation.
Figure 1:Circular genome map of Bacillus cereus sp. Strain ATCC 55673 with the first (outermost) and fourth rings depicting COG categories of protein coding genes on the forward and reverse strands, respectively.

Briefly, genomic DNA from the strain was extracted and prepared for sequencing on an Illumina HiSeq2000 instrument as described previously [15-19]. De novo assembly of the raw reads was performed with the SPAdes assembler [2] using default settings. Assembly coverage statistics were computed by mapping raw reads to the assembled genome using bowtie2 [12], contigs with coverage less than 37.65 were removed from the assembly. The remaining reads were aligned with nucmer [11] against the closest reference sequence from NCBI (determined by a BLAST of the 16S rRNA sequence): accession numbers CP002013.1, CP002014.1, and NC_014119.1 for chromosomes 1,2 and 3. All contigs aligned to these references, and the optimal contig ordering and orientation to most closely match the reference was determined using mummerplot [11] with layout specified. Contigs were then reordered and reversed as needed to match the ordering determined by mummerplot. The genome, with a coverage of 90x, was then annotated and genes predicted by IMG/er [14], RAST [1] and NCBI’s Prokaryotic Genomes Automatic Annotation Pipeline (PGAAP), version 2.0. A circular genomic map of strain VIA-1-2016 was generated using CG View [7] using the 70 assembled contigs with N50 contig length of 259,736 bases (Figure 1). The genomic size of strain ATCC 55673 was estimated to be approximately 460,08,79 bases with a G+C content of 37.45% Moreover, the strain contained a total of 4,590 putative genes, with 102 total RNA genes, 55 tRNA genes and 7 copies of the 16S rRNA genes, respectively.
Results
Genome-centric evaluation of strain ATCC 55673
Figure 2:Whole genome sequence based hierarchical clustering analysis phylotree of Bacillus cereus sp. strain ATCC 55673 relative to several other Bacillus spp.
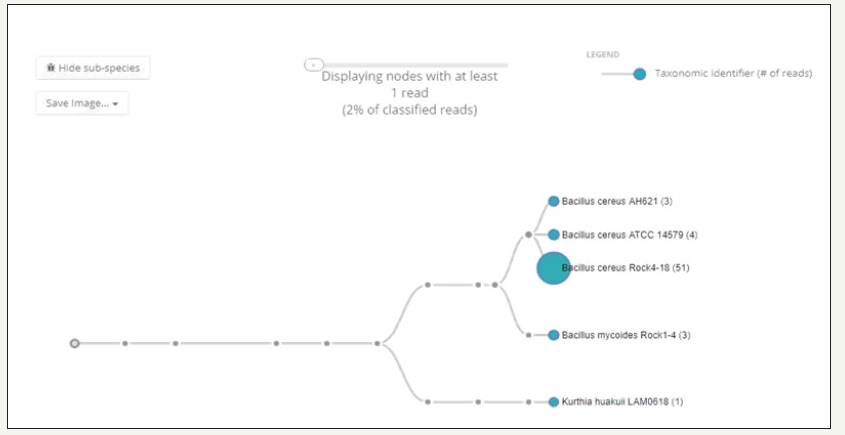
Genome-centric evaluation of strain ATCC 55673 revealed that out of 4,488 total protein coding genes, 65.34% genes associated with clusters of orthologous groups of proteins (COGs); 79.65% genes annotated to protein coding function predictions; 23.33% genes annotated as protein coding genes with enzyme production and 26.47% genes were connected to KEGG pathways. We then selected the whole genome sequences of 24 other Bacillus sp. strains that are available in IMG/er to run a comparative analysis of strain ATCC 55673 relative to other sequenced Bacillus strains, which revealed taxonomic affiliation with Bacillus cereus spp. CCGE1002 and H160, respectively (Figure 2). The Bacillus cereus group of beta-proteobacteria are ubiquitously distributed in diverse ecological habitats ranging from soil to aqueous environments, forming symbiotic associations with plants and animals, serving as saprophytes to endosymbionts and even as biocontrol agents of pathogens [5]. Moreover, a shared trait with many Bacillus sp. species are the presence of a large, multi-replicon genome with a plethora of multiple insertion sequences likely conferring genome plasticity and metabolic versatility to this genu [13]. It is interesting to note that the closest taxonomic relatives of strain ATCC 55673 were isolated from legume nodules [17] and shared approximately 190 CDS related to aromatic compound metabolism along with 150 common CDS for resistance to antibiotics and toxic compounds. Strain ATCC 55673 also affiliated closely with B. xenovorans LB400, which harbors one of the two largest known bacterial genomes [3]. Strain LB400 is a robust polychlorinated biphenyl-degrader and can also metabolize compounds containing single-carbon (C1) groups, isoflavonoids, diterpenoids, and sulfonates.
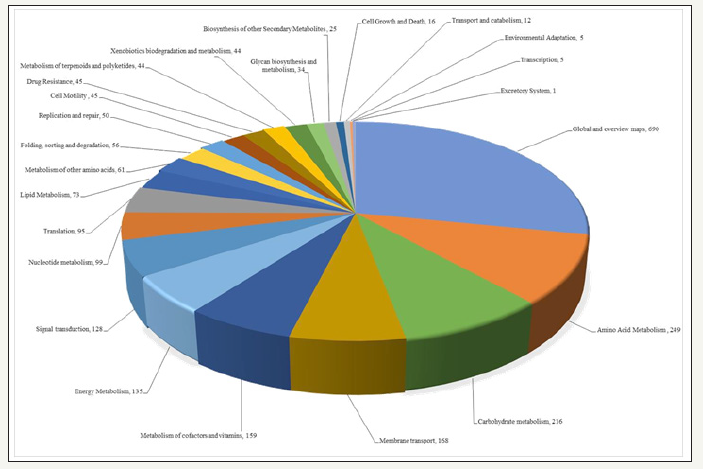
Furthermore, we performed a KEGG (Kyoto Encyclopedia of Genes and Genomes)-based hierarchical functional gene clustering analysis of ATCC 55673 (Figure 3), which revealed the presence of a total of 1215KEGG genes with the top 5 categories belonging to amino acid metabolism (222 genes); membrane transport (205 genes); carbohydrate metabolism (170 genes); energy metabolism, (122 genes), and metabolism of cofactors and vitamins (163 genes), respectively. Additionally, a total of 2,999 protein coding genes associated with COGs in strain ATCC 55673 were found with further classification into 24 categories. The following were most abundant COG subsystems in strain ATCC 55673: amino acid transport and metabolism (11.02%); transcription (8.86%); carbohydrate transport and metabolism (4.20%); energy production and conversion (4.63%); cell wall/membrane/envelope biogenesis (3.85%); lipid transport and metabolism (4.40%); signal transduction mechanisms (6.53%); inorganic ion transport and metabolism (7.54%); coenzyme transport and metabolism (5.58%); secondary metabolites biosynthesis, transport and catabolism (2.56%)-many of these features suggest genomic propensity of this strain to engage in high metabolic activity in its native contaminated environment. The strain also contains 7 biosynthetic gene clusters containing 169 genes; a total of 3.68% of its total genome. These analyses suggest Bacillus cereus strain ATCC 55673 to harbor various genome-enabled metabolic and catabolic processes that potentially facilitate its successful colonization and survival within the SRS co-contaminated soils.
Figure 3:Functional traits connected with KEGG pathways identified from the whole genome sequence of Bacillus cereus sp. strain ATCC 55673.
Discussion
Genes potentially conferring metal resistance in strain ATCC 55673
Figure 4:Shown is the chromosomal region of the cobalt/zinc/cadmium subsystem gene (1476bp) in Bacillus cereus sp. strain ATCC 55673relative to other bacteria. The graphic is centered on the focus gene, which is red and numbered 1. Sets of genes with similar sequence are grouped with the same number and color. Genes whose relative position is conserved in at least four other species are functionally coupled and share gray background boxes.
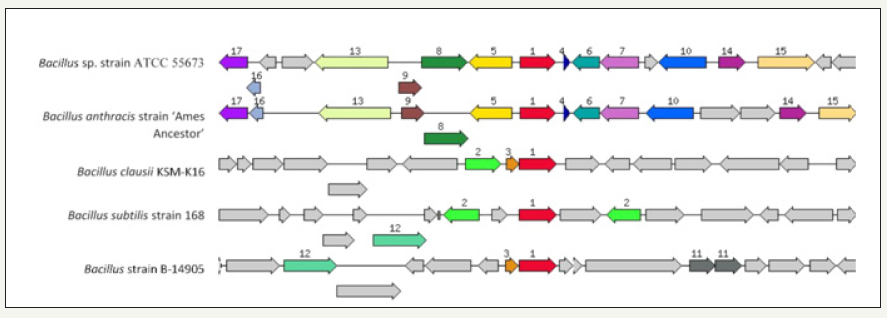
One notable attribute of the Bacillus genus is their vigorous ability to degrade xenobiotic compounds and resistance to various metals [17-22], including Al, As, Cd, Be, Cu, Cr, Fe, Pb, Ni, Se and Zn [8]. Genome-centric assessment of strain ATCC 5673 led to the identification of several gene homologues previously implicated in the resistance against heavy metals, such as efflux systems, metalloproteinase, transporters, permeases, and outer membrane factor (OMF) lipoproteins. More specifically, the cobalt/zinc/ cadmium resistant gene (CzcD) in strain ATCC 5673 was identified to be approximately 900bp in size (contig26007; |6666666.270904. peg.2402) which is shown relative to four similar organisms (Figure 4). We also identified the HoxN/HupN/NixA family high affinity nickel/cobalt transporter-permease gene complex (contig41300; fig|6666666.232708.peg.2084) which was 1029bp. In addition to these genes, bacterial resistance to metals is also governed by genes for the RND family proteins- these provide for resistance, nodulation, and cell division, respectively-all part of transenvelope protein complexes which detoxify the cellular environment by exporting toxic metal cations from the periplasm to the outside. Several RND-type efflux gene homologues were found interspersed within the genome of strain ATCC 55673 (data not shown). Moreover, the ABC-type transporters, heavy-metal-translocating P-type ATPases, and heavy-metal transport/detoxification proteins were plentifully represented in the genome of strain ATCC 55673, potentially maintaining metal homeostasis.
One mechanism used by Bacillus to colonize metals-rich environments, especially Fe, is by the re-oxidation of ferrous ion and generating various insoluble iron sulfides such as ferrous in sulfate, melanterite and coquimbite [6-9]. Therefore, such bacterial isolates can play a significant role in the remediation of metal contaminated waste. To identify the genomic basis for metal cycling by strain ATCC 55673, we manually queried the annotated genome revealing the presence of iron-sulfur cluster assembly ATPase protein SufC (contig59916; 6666666.270914.peg.3124 as well as iron-sulfur cluster assembly protein SufB (contig59916; 6666666.270914.peg.3120 and iron-sulfur cluster assembly protein SufD (contig59916; 6666666.270914.peg.3123 in strain ATCC 55673 (Figure 5).
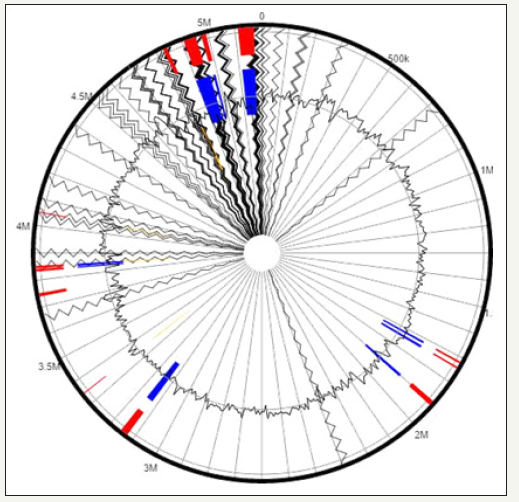
Figure 5:Putative genomic islands (GEIs) predicted within the genome of Bacillus cereus sp. strain ATCC 55673 when aligned against a reference Bacillus cereus sp. The outer black circle represents the scale line in Mbps and the black zig-zag line plot delineates each of the 175 contigs identified from strain SRS-W-2-2016. GEIs obtained from each of the following methods are shown in color: SIGI-HMM (orange), Island Path-DIMOB (blue), and integrated detection (red), respectively.
In addition to the metal related genes, several genes that potentially confer the ability to degrade organic contaminants were also identified in strain ATCC 55673, especially the oxygenases and oxidases such as multicopper oxidase (contig26007; 6666666.270914.peg.2529 (data not shown). This is relevant because oxygenase enzymes have been previously shown to cometabolize degradation of trichloroethylene (TCE), one of the widespread organic contaminants identified at the SRS site [9]. This potential shows that strain ATCC 55673 is metabolically active in the biomineralization of not only metals but also hydrocarbons in the coal pile run off at the Savannah River Site, including munitions wastes.
Genomic Islands (GEIs) in strain ATCC 55673
Another interesting genomic trait of strain ATCC 55673 are the presence of several Genomic Islands (GEIs) spread over the three chromosomes that are typical of this genus. Bacterial genomes consist of a core set of genes encoding for essential metabolic functions; these can be augmented with genes acquired from the bacterium’s native environment by horizontal gene transfer (HGT) mechanisms. The set of foreign genes recruited by the bacteria can provide the host with environmentally adaptive traits and genomic plasticity. Many such HGT-acquired genes occur as orthologous blocks commonly known as genomic islands (GEIs) [10] (Figure 5). GEIs have been more commonly known to render virulence or antibiotic resistance to the host bacteria but more recently, whole genome sequencing studies have also revealed other adaptive functional traits encoded by GEIs that are classified into the following 4 categories-pathogenicity islands (PAIs), that code for virulence genes; metabolic islands (MIs), genes for biosynthesis of secondary metabolites; resistance islands (RIs), genes that code for resistance- typically against antibiotics; and symbiotic islands (SIs), facilitating symbiotic associations of the host with other micro- and macro-organisms respectively. We used Island Viewer to identify genomic islands (GEIs) ATCC 55673, which predicts GEIs integrating two widely used sequence composition based GEI prediction methods- SIGI-HMM and Island Path-DIMOB along with a comparative GI prediction method- Island Pick. Interestingly, a plethora of genomic islands were identified in ATCC 55673 (Figure 6). When GEI-encoded homologues in strain ATCC 55673 were further analyzed by BLAST, several of them closely affiliated with genes previously implicated in the bioremediation of both metals and organic compounds, suggesting that GEIs were horizontally acquired by strain ATCC 55673 from other biodegradative bacteria to facilitate survival in its native environment that is cocontaminated by heavy metals, radionuclides, and aromatics.
Figure 6:Whole genome comparative alignment of Bacillus cereus strain ATCC 55673 with four close taxonomic relatives. Each of the genome sequence analyzed is presented horizontally with the scale showing the sequence coordinates and the conserved segments represented as the colored blocks which are connected across genomes.
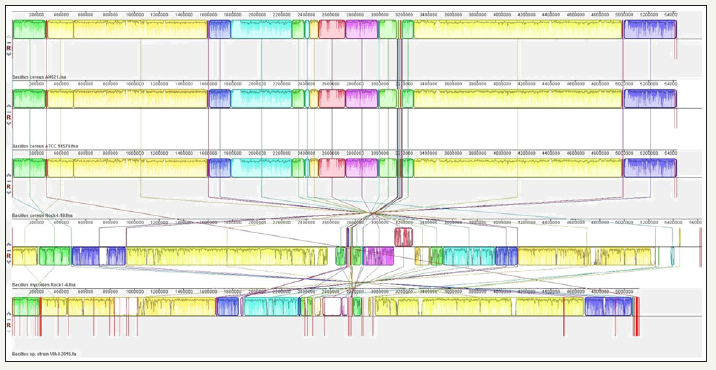
Genomic comparison of strain ATCC 55673
Figure 7:Whole genome comparative alignment of Bacillus cereus strain ATCC 55673 with closest taxonomic relative- B. cereus Rock 4.

Further comparative genomic analysis to identify the presence of large-scale evolutionary events, such as rearrangements and inversions, was performed by multiple alignment of strain ATCC 55673 which revealed the presence of several crisscrossing locally collinear blocks (LCBs), suggesting the complicated rearrangement history of strain ATCC 55673 relative to related Bacillus genomes. Regions that are inverted relative to strain ATCC 55673 appear shifted below a genome’s center axis (Figure 7). In fact, visual inspection of the genomic rearrangement pattern for s characteristic of symmetric inversion about the terminus, disti train ATCC 55673 relative to other Bacillus species revealed several instances of local overlapping inversions nguishable as a “fan” pattern of crossing lines. Therefore, we hypothesize that the genomic collinearity of strain ATCC 55673 was lost relative to its closest taxonomic relatives, most likely due to evolutionary mechanisms associated with genomic inversions and other rearrangements.
Figure 8:Growth dynamics of ATCC 55673 in 5, 10, 15 and 20ppm Uranium (A-D).
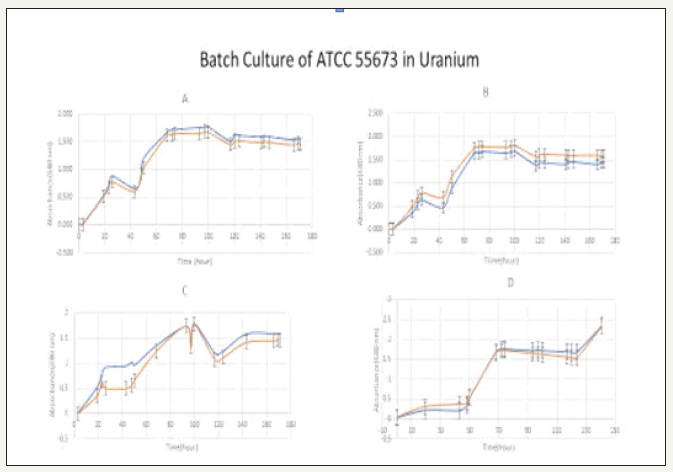
Genomic rearrangements of strain ATCC 55673 with the closest taxonomic relative, B. cereus Rock4 was confirmed by Nucmer analysis followed by visualization with circus (Figure 8). Genome alignment was performed using Nucmer with parameters -Maxgap 5000 and -b 500. Contigs were ordered by length, and alignments of at least 5kb were visualized using circoA. Same-stranded alignments were plotted with red ribbons, and reverse-stranded alignments with blue ribbons.
Batch culture experiments of ATCC 55673 in uranium biomineralization
Soil samples were collected from SRS according to Seaman et al., 2005. Briefly, field-moist sediments collected from site 101 along the Tims Branch at SRS were collected, homogenized and sieved (2mm) to remove stones and large organic debris i.e. stems, roots, and leaves. Soil aggregates were gently crushed by hand to pass through the sieve. Batch equilibrations generally followed the methods outlined in Arey et al. (1999) and Seaman et al. (2001a, 2001b, 2003).
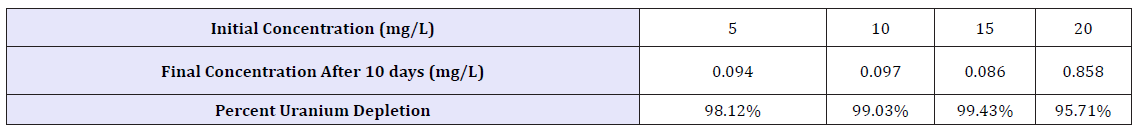
To demonstrate tolerance and biomineralization of ATCC 55673 in these uranium-contaminated soils, samples were first autoclaved and made into slurry using Artificial Groundwater (AW). In these experiments, 495mL of AW +5g of autoclaved soil, infused with LB were serially diluted and plated out. ATCC 55673 was streaked and grown overnight at 35 °C. Colony forming units (CFUs) were determined for each plate (Figure 8). In other batch experiments, cultures were grown in flasks with additional spikes of 5 -20mg/L of Uranium. Depletion of uranium was determined using Inductively Coupled Plasma Mass Optical Emission Spectrometry (Perkin Elmer). Percent uranium depletion ranged above 90% in all concentrations (Table 2), indicating biomineralization and tolerance of ATCC 55673 in uranium.
Table 2:Biomineralization of Uranium ATCC 55673.
Conclusion
Genome-centric analysis of metals and various dissolved metals resistant microbiota isolated from waste water sludge and then tested on coal pile runoff water from historically contaminated environments such as the Savannah River Site (SRS), are lacking. SRS is a former nuclear weapons production facility located near Aiken, SC which is co-contaminated with heavy metals, radionuclides and Volatile Organic Compounds (VOCs). Moreover, ongoing contamination at the SRS has also been linked the coal fired plant at this site which releases metal contaminated wastewater into a catchment basin especially during rainfall events. Because environmental microorganisms can underpin metal biotransformation including radionuclide precipitation, sorption, intracellular accumulation and biomineralization, this study is a genome-centric assessment to probe for those genes that facilitate bioremediation of environmental contaminants specific to the SRSs. Specifically, this study facilitates a deeper understanding of dissolved metals and other metals resistance of a Bacillus cereus sp. strain ATCC 55673 isolated from an activated sludge habitat. Overall, we show that Bacillus cereus sp. strain ATCC 55673, which was isolated on very high concentrations of various metals such as As, Al, Fe, Cu, Cr, Pb, and Zn possesses a suite of ecologically relevant genomic traits against contaminants to include substrate binding proteins, permeases, transport regulators and efflux pumps all targeted to continuously shunt metals to the extra-cellular environment or prevent their cellular uptake so the bacterium can persist along with the mixed contaminants. It also appears that horizontal gene transfer is an evolutionary mechanism that equips microbiota to facilitate their colonization and proliferation of radionuclide and hydrocarbon contaminated environments. Studies such as this will provide a better understanding on bioremediation for rehabilitation and stewardship of historically polluted environments.
Acknowledgement
This work was supported by the Department of Energy Minority Serving Institution Partnership Program (MSIPP) managed by the Savannah River National Laboratory under SRNS contract DEAC09- 08SR22470.
Nucleotide Sequence Accession Number
The Whole Genome Shotgun project of Bacillus cereus sp. strain ATCC 55673 reported in this study has been deposited at DDBJ/ ENA/Gen Bank under the accession #MSDV00000000 (www.ncbi. nlm.nih.gov/nuccore/MSDV00000000).
References
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, et al. (2008) The RAST server: Rapid annotations using subsystems technology. BMC Genomics 9: 75.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. (2012) SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19(5): 455-477.
- Chain PS, Denef VJ, Konstantinidis KT, Vergez LM, Agulló L, et al. (2006) Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc Natl Acad Sci 103(42): 15280-15287.
- Chauhan A, Pathak A, Ewida AY, Griffiths Z, Stothard P (2016) Whole genome sequence analysis of an Alachlor and Endosulfan degrading Pseudomonas strain W15Feb9B isolated from Ochlockonee River, Florida. Genom Data 8: 134-138.
- Coenye T, Vandamme P (2003) Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ Microbiol 5(9): 719-729.
- Dhillon BK, Laird MR, Shay JA, Winsor GL, Lo R, et al. (2015) Island viewer 3: More flexible, interactive Genomic Island discovery, visualization and analysis. Nucleic Acids Res 43(W1): W104-W108.
- Grant JR, Arantes AS, Stothard P (2012) Comparing thousands of circular genomes using the CG view comparison tool. BMC Genomics 13: 202.
- Ibeanusi, V, Phinney D, Thompson M (2003) Removal and recovery of metals from a coal pile runoff. Environmental Monitoring and Assessment 84(1-2): 35-44.
- Jackson D (2001) Characterization activities to evaluate chlorinated solvent discharges to Tims Branch from the A/M area of the Savannah River Site. Savannah River Site, USA.
- Juhas M, Van der Meer JR, Gaillard M, Harding RM, Hood DW, et al. (2009) Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev 33(2): 376-393.
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, et al. (2004) Versatile and open software for comparing large genomes. Genome Biol 5(2): R12.
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357-359.
- Lessie TG, Hendrickson W, Manning BD, Devereux R (1996) Genomic complexity and plasticity of Burkholderia cepacian. FEMS Microbiol Lett 144(2-3): 117-128.
- Markowitz VM, Mavromatis K, Ivanova NN, Chen IMA, Chu K, et al. (2009) IMG/ER: A system for microbial genome annotation expert review and curation. Bioinform 25(17): 2271-2278.
- (1999) Nuclear Regulatory Commission.
- (2000) Nuclear Regulatory Commission.
- Ormeño OE, Rogel MA, Chueire LMO, Tiedje JM, Martínez RE, et al. (2012) Genome sequences of Burkholderia sp. strains CCGE1002 and H160, isolated from legume nodules in Mexico and Brazil. J Bacteriol 194(24): 6927-6927.
- Oyetibo GO, Ilori MO, Obayori OS, Amund OO (2013) Biodegradation of petroleum hydrocarbons in the presence of nickel and cobalt. J Basic Microbiol 53(11): 917-927.
- Pathak A, Chauhan A, Ewida AY, Stothard P (2016) Whole genome sequence analysis of an Alachlor and Endosulfan degrading Micrococcus sp. strain 2385 isolated from Ochlockonee River, Florida. J Genomics 4: 42-47.
- Pantoja PD, Nikel PI, Chavarría M, De Lorenzo V (2013) Endogenous stress caused by faulty oxidation reactions fosters evolution of 2, 4-dinitrotoluene-degrading bacteria. PLoS Genete 9(8): e1003764.
- Suárez MZR, Caballero MJ, Coutinho BG, Mendonça PL, James EK, et al. (2012) Common features of environmental and potentially beneficial plant-associated Burkholderia. Microb Ecol 63(2): 249-266.
- Yang Z, Zhang Z, Chai L, Wang Y, Liu Y, et al. (2016) Bioleaching remediation of heavy metal-contaminated soils using Burkholderia sp. Z-90. J Hazard Mater 301: 145-152.
© 2018 Victor Ibeanusi,Ashish Pathak. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






