- Submissions

Full Text
Significances of Bioengineering & Biosciences
Cpg Methylation in G-Quadruplex and IMotif DNA Structures
Isaakova E, Varizhuk A and Pozmogova G*
Research and Clinical Center of Physical-Chemical Medicine, Moscow, Russia
*Corresponding author: Pozmogova G, Research and Clinical Center of Physical-Chemical Medicine, Moscow, Russia
Submission: March 14, 2018;Published: April 04, 2018

ISSN 2637-8078Volume1 Issue3
Abstract
Abberant hypomethylation in DNA regions with noncanonical folding potential (ncDNA motifs) is believed to predetermine tumor development - presumably, by facilitating G-quadruplex (G4) and/or i-motif (IM) formation via altering nucleosome positioning (stable G4s induce subsequent genomic rearrangements). We questioned whether CpG methylation per se affects the dsDNA-ncDNA equilibrium. Thermodynamic studies of genomic and model oligonucleotides with methylated CpG sites at different positions are reported. The genomic oligonucleotides analyzed in this work are DNA fragments with reportedly different methylation statuses in colorectal cancer and normal cells. Free energies of duplex, ncDNA formation from single strands were calculated based on melting curve analyses. Polyethylenglycole was used to imitate crowding effect. Our results suggest that CpG methylation may alter the energetic barrier for dsDNA-IM transitions.
Keywords: DNA secondary structures; Methylation; G-quadruplexes; i-motifs; Thermodynamic stability; Molecular crowding
Abbreviations: CD: Circular Dichroism; CGI: Cpg Islands; Etbr: Ethidium Bromide; G4: G-Quadruplex; HPLC: High Performance Liquid Chromatography; IM: Intercalated Motif (I-Motif); MALDI-TOF; MS: Matrix Assisted Laser Desorption Ionization: Time of Flight Mass Spectrometry; Ncdna: Noncanonical DNA Structures; ON: Oligonucleotide; PEG: Polyethylene Glycol; RRT: Rotational Relaxation Time; TDS: Thermal Difference Spectrum
Introduction
DNA-methylation in mammalian somatic cells occurs predominantly due to cytosine modification in CpG dinucleotides. The methylation pattern distinguishes two different fractions of CpG composition: the main fraction in which CpG dinucleotides are infrequent (on average, 1 CpG per 100 b.p.), but heavily methylated; and the secondary fraction, known as CpG islands (CGIs), which is represented by short DNA fragments (~ 1000 bp) with average CpG frequency of 1 per 10 b.p. [1]. Most CGIs contain sites of transcription initiation, which are typically demethylated (i.e., contain no methyl groups at the C5 position of cytosine residues) in an otherwise heavily methylated genome, even when the corresponding gene is transcriptionally inactive. There are, however, examples of promoter CGIs that become methylated, leading to stable silencing of the respective genes. Both hypermethylation of promoter CGIs (i.e., the excess of 5-methylcytosine residues) and genome-wide hypomethylation (the loss of the methyl groups in normally methylated CpG sites) are regarded as hallmarks of cancer genomes [2-4]. In this work we describe impacts of colorectal cancerassociated aberrant methylation on the propensity of the respective DNA fragments for conformational rearrangements.
One possible mechanism of aberrant hypomethylation-induced oncogenesis has been proposed previously based on the analysis of genomic distributions of noncanonical secondary structures (ncDNA) and DNA breakpoints associated with somatic copy number alterations in cancer tissues [5]. The proposed mechanism includes the following steps: (i) random epigenetic mutation changes tissue-specific methylation pattern; (ii) hypomethylation adjacent to ncDNA motifs favors ncDNA folding in the presence of stabilizing proteins and negative supercoiling; (iii) nсDNA increases the likelihood of further alterations (DNA damage and genomic rearrangements).
Four-stranded noncanonical structures of G/C-rich DNA, i.e., G-quadruplexes (G4s – planar arrangements of guanine tetrads) and i-motifs (IMs - intercalated parallel duplexes stabilized by hemiprotonated C-C pairs) have recently gained significant attention as possible regulatory elements and the hotspots of genomic or epigenetic instability [6-9]. Although IMs are typically stable at pH 3-6.5, recent data support possibility of IM formation under physiological conditions [10]. Both G4s and IMs can repress or activate gene expression at the transcriptional level, which is, in some cases, beneficial for genome maintenance. For instance, C-myc expression is presumably controlled by the interplay between protein factors recognizing mutually exclusive G4 and IM sites in the promoter region of this oncogene [11].
Methylation effects on folding and thermal stability of G4 motifs colocalized with CGIs in promoter regions of several genes have been investigated previously [12,13]. The epigenetic modification was shown to alter G4 topology and thermal stability. However, the data are somewhat inconclusive and no general tendencies can be outlined. For instance, methylation reportedly stabilized the G4 structure in the BCL-2 promoter, but had the opposite effect on the MEST promoter G4. There is also some controversy about IMs: single and double Me-CpG insertions have been shown to enhance thermal stabilities of telomeric IM structures, while further methylation caused destabilization [14].
The influence of CpG methylation in G4 motifs or the opposing IMs on the ‘B-DNA–ncDNA’ equilibrium remains unclear at the moment. In this paper, we report the comparative analysis of ncDNA and B-DNA thermodynamic stabilities in methylated and demethylated states. The impacts of methylation rate (single or several Me-CpG sites within the same G4 / IM) and Me-CpG positions, as well as molecular crowding effects, are discussed.
Our major goal was to clarify the dependence of DNA refolding potential on methylation status, which may shed new light on the role of epigenetic alterations in oncogenesis.
Materials and Methods
Oligonucelotide synthesis, purification and MS analysis
Oligonucleotides (ONs) were synthesized on a ASM-800 DNA synthesizer (Biosset, Russia) following standard phosphoramidite protocols using standard reagents. For methylated ONs, we additionally used 5-Mе dC CE phosphoramidite (Glen Research, USA). The ONs were purified by preparative scale reverse-phase HPLC as described in [15] (for IMs) and [16] (for G4s). The final purity of all ONs was determined to be 95% by HPLC. MALDI-TOF MS analysis of the ONs was performed as described in [16]; the data were acquired on a Microflex mass spectrometer (Bruker, USA).
UV absorption and circular dichroism spectroscopy
UV absorption and circular dichroism (CD) spectra were recorded on a Chirascan spectrophotometer (Applied Photophysics, UK), equipped with a thermostated cuvette holder. Solutions of G4- or IM-forming ONs (2.5μM) in 20mM Tris-HCl buffer (pH 7.6 or 5.5, respectively) containing 100mМ KCl were annealed rapidly (heated to 95 °C and snap-cooled on ice) prior to measurements to facilitate intramolecular folding. Molar CD per nucleotide residue was calculated as follows: Δε = θ/(32.982 × C × l × n), where θ is ellipticity (degree), C is ON concentration (M); l is optical pathlength (cm) and n is the number of nucleotide residues in the ON. Thermal difference spectra (TDS) were obtained by subtracting absorption spectra registered at 15 °C from the spectra registered at 90 °C.
Rotational relaxation time assay
Rotational relaxation times (RRT) of the fluorescent intercalator – ethidium bromide (EtBr) in complexes with G4s/ IMs (1:1) were calculated based on EtBr fluorescence polarization and lifetime values using the PerrineWeber equation [17,18]. The latter values were estimated as described in [19] using Cary Eclipse spectrophotometer (Agilent technologies, USA) and Easy Life V fluorescence lifetime fluorometer (Horiba, Japan).
Melting experiments
In UV-melting experiments, absorption at 295nm (for G4 and IMs) or 260nm (for duplexes) was registered from 5 °C to 90 °C every 1 °C at a heating rate of 0.5 °C/min. Polyethylene glycol with average molecular weight of 200 (PEG 200) was added to ON solutions to a final concentration of 40% to imitate molecular crowding conditions. All G4 and IM samples were annealed rapidly prior to the experiments as described in the previous subsection. Duplex samples were heated to 95 °C and cooled slowly to room temperature.
The melting curves were analyzed and fitted using DataFit9 software (Oakdale Engineering, USA).
Results and Discussion
Oligonucleotide design
In this study we analyzed model sequences and fragments of the human genome that are differentially methylated in colorectal cancer and normal tissues. The latter sequences were selected based on previously published results of genome-scale methylation profiling with Infinium HumanMethylation450 chips [20]. The coordinates of CpG sites with replicable and statistically significant differential methylation (candidate tumor markers) were overlaid with the coordinates of putative G4/IM sites obtained using ImGQFinder software [21]. Secondary structures of the presumed G4s and IMs containing candidate CpG/Me-CpG tumor markers were verified by optical methods, and the structures with ambiguous folding were excluded from subsequent analysis.
Sequences of the selected genomic fragments (methylated and demethylated variants), as well as model ONs, are provided in Table 1. G4 oplah1 is a fragment of the 5-oxoprolinase gene OPLAH; IMs Lmo1 and Fli1 are fragments the respective transcription factorcoding genes, and ppp is a fragment of the protein phosphatase 1 gene PPP1R16. Model ONs Ib (IM), mbb (G4) and their derivatives were designed to contain methylation sites at different positions: in G4/IM loops, at loop/G-tetrad boundaries or inside the IM core (Me-C in hemiprotonated C-C pairs).
Secondary structure verification
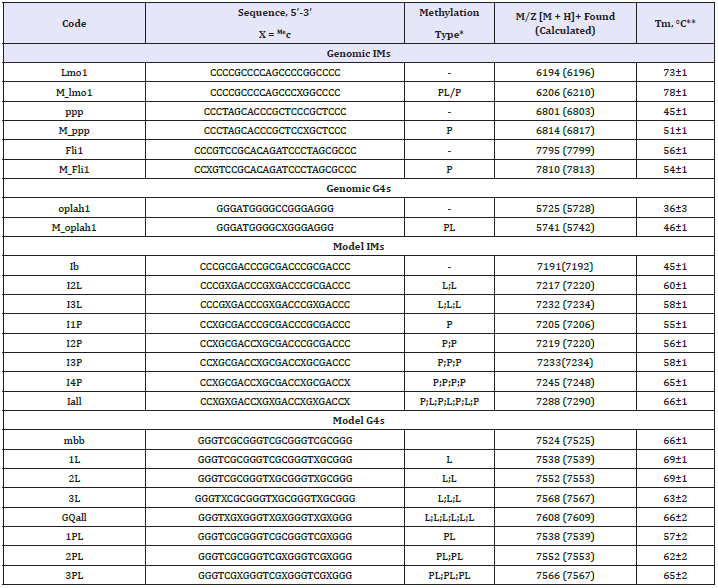
The G4 and IM structures were characterized by optical methods. The results are summarized in Figures 1 and 2 for genomic and model ONs, respectively.
Thermal difference and circular dichroism spectra (TDS and CD) were used to confirm G4/IM formation. Rotational relaxation time (RRT) assay with EtBr as a fluorescent intercalator was used to distinguish between inter- and intramolecular structures (RRT is roughly proportional to the hydrodynamic volume of the intercalator/ON complex). UV melting experiments were performed to assess thermal stabilities of the structures, and the melting temperatures (Tm) are provided in Table 1.
Table 1: Oligonucleotide sequences, methylation positions, MS data and G4/IM melting temperatures.
*P, MeC in C-C pairs; L, MeC in IM/G4 loops; PL, MeC at loop/G-tetrad boundaries.
**In 20 mM Tris-HCl buffer (pH 7.6 and 5.5 for G4s and IMs, respectively) containing 100 mM KCl.
Figure 1: Characterization of genomic methylated and demethylated G4 (A) and IMs (B) by optical methods. Circular dichroism spectra (CD per mole of nt.) at 15 °C, thermal difference spectra (TDS), rotational relaxation times (RRT) of EtBr in complexes with G4s/IMs and G4/IM UV-melting curves are shown. RRT control is a DNA duplex TCACCTCCCTCC/GGAGGGAGGTGA (24 nt. residues in total). Conditions: 2.5μM ON, 20 mM Tris-HCl (pH 7.6 and 5.5 for G4s and IMs, respectively) and 100mM KCl.
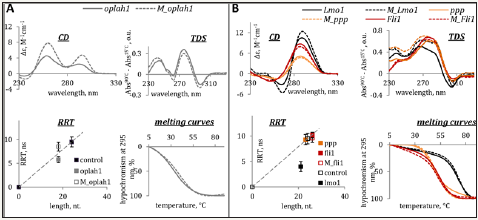
Figure 2: Characterization of model G4 (A) and IMs (B) by optical methods. Circular dichroism spectra (CD per mole of nt.) at 15 °C, thermal difference spectra (TDS), rotational relaxation times (RRT) of EtBr in complexes with G4s/IMs and G4/IM UV-melting curves are shown. RRT control is a DNA duplex TCACCTCCCTCC/GGAGGGAGGTGA (24 nt. residues in total). Conditions: 2.5μM ON, 20mM Tris-HCl (pH 7.6 and 5.5 for G4s and IMs, respectively) and 100mM KCl.

CD spectra and TDS of oplah1 (top panels in Figure 1A) and mbb (top panels in Figure 2A) contain specific signatures of G4 DNA [22,23]. Positive CD bands at 265 and 295 nm point to hybrid G4 topologies. RRTs of EtBr in complexes with the ONs (left bottom panels in the figures) suggest intramolecular folding. Melting profiles (right bottom panels) indicate moderate and high thermal stabilities of oplah1 and mbb, respectively.
ONs ppp, Fli1, Lmo1 and Ib adopt thermodynamically stable intramolecular IMs, as evident from Figure 1B and Figure 2B. Characteristic features of IM structures are present in the CD spectra (positive band at 285nm) and TDS (negative band at 295nm); RRT values are close to the calibration line (this implies intramolecular folding) in all cases except lmo1. The EtBr/lmo1 RRT value is reduced - presumably, due to a particularly compact folding of the lmo1 IM.
Methylation effects on thermal stabilities of the secondary structures
Methylation of the genomic G4 oplah1 at the loop/tetrad boundary enhanced thermal stability of the structure, as evident from the somewhat shifted melting curve and increased CD amplitude of M_oplah1 (ΔTm ≈ +10 °C). The general shapes of oplah1 and M_oplah1 CD spectra are rather similar, suggesting no principal changes in the G4 topology (Figure 1A).
In the case of the model G4 mbb, both loop CpG methylation and loop/tetrad CpG methylation had minor effects on Tm values, while the portion of the folded structure appears to decrease with increasing methylation rate (see CD amplitudes and hyper/ hypochromism changes in Figure 2A). The latter effect is particularly pronounced for Me-CpG sites at loop/tetrad boundaries. The extensively methylated G4 GQall seems to be mostly unfolded (the characteristic negative TDS band at 295 nm is absent) despite the relatively high apparent Tm value. Thr increase of RRT values with increasing methylation rate can be attributed to the fact that methylation per se contributes to the G4 hydrodynamic volume. However, partial aggregation, i.e., intermolecular (possibly non-G4) folding of heavily methylated mbb derivatives cannot be excluded.
As concerns IMs, methylation had moderate (mostly stabilizing) effects on the genomic structures (Figure 1B) and profound stabilizing effects on the model structure (Figure 2B) for both core and loop cytosine modifications (ΔTm ≈ +10 °C for a single Me-C in the core; further modification had less pronounced effects). Overall, the results obtained for the IM structures agree with the data in the literature, except that no negative impact of extensive methylation [14] was observed. Subsequent in-depth analysis was performed for IMs exclusively.
Molecular crowding effects
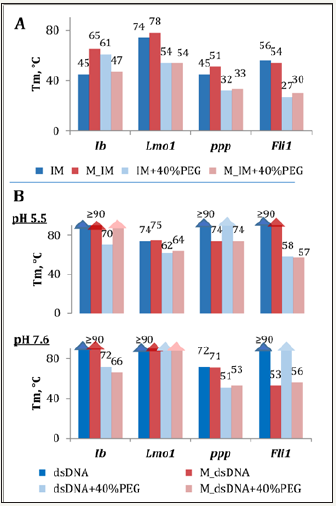
We questioned whether substantial influence of CpG methylation on IM thermal stabilities is maintained under molecular crowding conditions. Polyethylene glycol of relatively a low average molecular weight (PEG 200) was used as crowding agent (higher molecular weight PEG is known to form nonspecific complexes with DNA, which may interfere with IM folding [24]). As evident from Figure 3A, PEG 200 tends to mitigate (Lmo1 and ppp) or even reverse (Ib and Fli1) methylation effects in IMs. Genomic IMs (both methylated and demethylated) exhibited decreased Tm values in the presence of PEG, and the opposite effect was observed for model structures.
According to the data in the literature, crowding usually has positive impacts IM thermal stabilities at near-physiological pH [24,25]. Stabilization is sometimes attributed to general factors, such as excluded volume (compact structures are more favorable in crowded microenvironment [26,27]). A more specific interpretation implies strengthened Cyt+/Cyt interactions due to the lowered solvent dielectric constant [24]. At pH values close to or substantially below IM transition value (pH, at which 50% of the ON strands are folded) the effect can be reversed – presumably, due to strengthened repulsion between opposing Cyt+ residues. Thus, different sensitivities of our model and genomic IMs to the crowding agent may be accounted for by different pH sensitivities.
Comparative analysis of B-DNA and i-motif stabilities
To clarify whether methylation may shift the B-DNA-ncDNA equilibrium, we compared thermal stabilities of IMs and respective duplexes for both methylated and demethylated variants. To obtain duplexes, the IM mixtures with complementary strands (1:1) were annealed slowly, and CD spectra were recorded to verify B-DNA folding. Spectra of the duplexes contained B-DNA signatures rather than IM/G4 signatures or their superposition at both acidic and neutral pH. Notable exceptions included Lmo1 and its methylated analog M_Lmo1 (under acidic non-crowded conditions these remarkably stable IMs appear to dominate even in the presence of the complementary strand).
All duplexes (demethylated variants in particular) displayed high thermal stabilities under non-crowded conditions (Figure 3B). PEG destabilized the duplexes in most cases, which is in line with the previously published data [26]. Predictably, duplexes are in general thermodynamically more favorable that IMs even at pH 5.5 irrespective of the methylation status. However, methylation alters the Tm ratio and free energies of secondary structure formation from single strands. This implies a possible impact on activation energy of the B-DNA↔ncDNA rearrangements. We addressed this matter for the model ON Ib and its extensively methylated analog I4P using a simplified approach (possible intermediates other than ssDNA were not taken into account).
Figure 3: Effects of molecular crowding and methylation on thermal stabilities of IMs (A) and respective duplexes (B). The IM samples were annealed rapidly at pH 5.5 prior to melting experiments; the dsDNA samples were annealed slowly at pH 5.5 or 7.6. M_IM – methylated IM (IP4 in the Ib series); M_dsDNA – duplex composed of M_IM and a complementary strand.
Enthalpic and entropic contributions to free energies of IM or dsDNA formation from single strands were obtained from the melting curves using and a two-state model [28], and the free energies were normalized to the physiological temperature. The results are summarized in Figure 4. Methylation of Ib increased the energetic barrier for the IM→duplex transition from 0.6 to 2.5kcal/ mol at 37 °C in non-crowded microenvironment (left panels in Figure 4). The effects were reversed in the presence of PEG: methylation lowered the IM→duplex barrier from 1.8 to 0.6 kcal/ mol and prohibited reverse transitions (right panels in Figure 4).
Figure 4: Methylation effects on dsDNA–ncDNA transition barriers (free energy diagrams). In each diagram, dashed lines (ssDNA) denote zero energy level, and the vertical arrows illustrate free energies of dsDNA (left) or IM (right) formation from single strands. Demethylated DNA under non-crowded conditions, dark blue; demethylated DNA under crowded conditions, light blue; methylated DNA under non-crowded conditions, dark pink; methylated DNA under crowded conditions, light pink.

Conclusion
We have shown that CpG methylation has diverse and in some cases pronounced effects on thermal stabilities of ncDNA structures and ncDNA-dsDNA transition probabilities. Substantial stabilization upon single-point methylation was demonstrated for a G4 structure and two IM structures representing genomic fragments with differential methylation in normal and colorectal cancer cells, but the crowding agent mitigated those stabilizing effects. Extensive methylation of the model IM structure caused dramatic stabilization. Importantly, the effect was reversed in crowded microenvironment.
Our findings support the hypothesis of methylation-induced conformational rearrangements of genomic DNA and illustrate the importance of accounting for crowding effects.
Acknowledgements
This work was supported by Russian Science Foundation [14- 25-00013].
References
- Deaton AM, Bird A (2011) CpG islands and the regulation of transcription. Genes Dev 25: 1010-1022.
- Szyf M (2008) The role of DNA hypermethylation and demethylation in cancer and cancer therapy. Curr Oncol 15(2): 72-75.
- Cheung HH, Lee TL, Rennert OM, Chan WY (2009) DNA methylation of cancer genome. Birth Defects Res C Embryo Today 87: 335-350.
- Hon GC, Hawkins RD, Caballero OL, Lo C, Lister R, et al. (2012) Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res 22(2): 246-258.
- De S, Michor F (2011) DNA secondary structures and epigenetic determinants of cancer genome evolution. Nat Struct Mol Biol 18 (8): 950-955.
- Benabou S, Avino A, Eritja R, Gonzalez C, Gargallo R (2014) Fundamental aspects of the nucleic acid i-motif structures. Rsc Advances 4: 26956- 26980.
- Murat P, S Balasubramanian. (2014) Existence and consequences of G-quadruplex structures in DNA. Current Opinion in Genetics & Development 25: 22-29.
- Cea V, Cipolla L, Sabbioneda S (2015) Replication of Structured DNA and its implication in epigenetic stability. Front Genet 6: 209.
- Katapadi VK, Nambiar M, Raghavan SC (2012) Potential G-quadruplex formation at breakpoint regions of chromosomal translocations in cancer may explain their fragility. Genomics 100(2): 72-80.
- Wright EP, Huppert JL, Waller ZAE (2017) Identification of multiple genomic DNA sequences which form i-motif structures at neutral pH (vol 45, pg 2951, 2017). Nucleic Acids Res 45(6): 2951-2959.
- Sutherland C, YX Cui, HB Mao, LH Hurley (2016) A Mechanosensor Mechanism Controls the G-Quadruplex/i-Motif Molecular Switch in the MYC Promoter NHE III1. Journal of the American Chemical Society 138: 14138-14151.
- Lin J, Hou JQ, Xiang HD, Yan YY, Gu YC, et al. (2013) Stabilization of G-quadruplex DNA by C-5-methyl-cytosine in bcl-2 promoter: Implications for epigenetic regulation. Biochemical and Biophysical Research Communications 433(4): 368-373.
- Stevens AJ, Kennedy MA (2017) Structural Analysis of G-Quadruplex Formation at the Human MEST Promoter. PLoS ONE 12:
- Xu BC, Devi G, Shao FW (2015) Regulation of telomeric i-motif stability by 5-methylcytosine and 5-hydroxymethylcytosine modification. Organic & Biomolecular Chemistry 13: 5646-5651.
- Protopopova AD, Tsvetkov VB, Varizhuk AM, Barinov NA, Podgorsky VV, et al. (2018) The structural diversity of C-rich DNA aggregates: unusual self-assembly of beetle-like nanostructures. Physical Chemistry Chemical Physics 20: 3543-3553.
- Prokofjeva M, Tsvetkov V, Basmanov D, Varizhuk A, Lagarkova M, et al. (2017) Anti-HIV Activities of Intramolecular G4 and Non-G4 Oligonucleotides. Nucleic Acid Therapeutics 27: 56.
- Weber G, Anderson SR (1969) The effects of energy transfer and rotational diffusion upon the fluorescence polarization of macromolecules. Biochemistry 8: 361-371.
- Beschetnova IA, Kaluzhny DN, Livshits MA, Shchyolkina AK, Borisova OF (2003) Ethidium probing of the parallel double- and four-stranded structures formed by the telomeric DNA sequences dG(GT)4G and d(GT)5. Journal of Biomolecular Structure & Dynamics 20: 789-799.
- Vlasenok M, Varizhuk A, Kaluzhny D, Smirnov I, Pozmogova G (2017) Data on secondary structures and ligand interactions of G-rich oligonucleotides that defy the classical formula for G4 motifs. Data in Brief 11: 258-265.
- Naumov VA, Generozov EV, Zaharjevskaya NB, Matushkina DS, Larin AK, et al. (2013) Genome-scale analysis of DNA methylation in colorectal cancer using Infinium HumanMethylation450 BeadChips. Epigenetics 8: 921-934.
- Varizhuk A, Ischenko D, Tsvetkov V, Novikov R, Kulemin N, et al. (2017) The expanding repertoire of G4 DNA structures. Biochimie 135: 54-62.
- Mergny JL, Li J, Lacroix L, Amrane S, Chaires JB (2005) Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic Acids Res 33: e138.
- Kypr J, Kejnovska I, Renciuk D, Vorlickova M (2009) Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res 37(6): 1713-1725.
- Cui J, Waltman P, Le VH, Lewis EA (2013) The effect of molecular crowding on the stability of human c-MYC promoter sequence I-motif at neutral pH. Molecules 18(10): 12751-12767.
- Rajendran A, Nakano S, Sugimoto N (2010) Molecular crowding of the cosolutes induces an intramolecular i-motif structure of triplet repeat DNA oligomers at neutral pH. Chem Commun (Camb) 46(8): 1299-1301.
- Nakano S, Miyoshi D, Sugimoto N (2014) Effects of molecular crowding on the structures, interactions, and functions of nucleic acids. Chemical reviews 114(5): 2733-2758.
- Kilburn D, Roh JH, Guo L, Briber RM, Woodson SA (2010) Molecular crowding stabilizes folded RNA structure by the excluded volume effect. J Am Chem Soc 132(25): 8690-8696.
- Marky LA, Breslauer KJ (1987) Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers 26(9): 1601-1620.
© 2018 Pozmogova G. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






