- Submissions

Full Text
Research in Pediatrics & Neonatology
Differentiation between C. glabrata and C. parapsilosis Isolated from Children with Diarrhea by PCR Amplification of the ITS1-5.8S-ITS2 rDNA
Ali R Hameed1, Sabah M Ali2 and Luma T Ahmed1
1Department of Microbiology, University of Diyala, Iraq
2Department of Pediatrics, Al-Al-Mustansiriyah University Baghdad, Iraq
*Corresponding author: Sabah M Ali, Research Scholar, Department of Pediatrics, College of Medicine, Al-Al-Mustansiriyah University Baghdad , Iraq, Email: sabahma2002@yahoo.com
Submission: February 27, 2018;Published: April 09, 2018

ISSN: 2576-9200 Volume2 Issue1
Abstract
Candida glabrata and Candida parapsilosis are most important non-albicans species which are considered pathogenic to human and isolated from children with diarrhea. The purpose of the study is the identification and differentiation between Candida glabrata and Candida parapsilosis among children with diarrhea by PCR and determination relation of this species with gender, age group and resident of children. Hundred stool samples were collected from children with diarrhea seeking health care at AL-Batool Hospital in Baqubah city. The Stool cultured on sabouraud dextrose agar, subcultured on Chromogenic agar and PCR methods identification. In the present study, the isolated were identified which 14/64 (21.9%) isolates were C. glabrata, while 11/64 (17.2%) isolate C. parapsilosis and 39/64 (60.9%) isolates were other Candida spp, the results showed there is a significant relationship between C. glabrata and C. parapsilosis with the gender of children. Candida glabrata and Candida parapsilosis have a close color between of them in Chromogenic agar but when PCR amplifying the ITS1-5.8S-ITS2rDNA region produced different size base pair that makes it easy differentiation between them.
Keywords: C. glabrata; C. parapsilosis; Diarrhea; ITS1-5.8S-ITS2rDNA; PCR
Introduction
Candida glabrata and Candida parapsilosis, the two genus belongs to the kingdom Fungi, division Eumycota (true fungi), which relates to class Deuteromycetes (Fungi imperfection) and the family Saccharomycetaceae (budding yeast) containing different genera of yeast, the genes of Candida which is one of the most common yeasts.
Candida glabrata constituting number two in greatest virulent yeast following C. albicans. C. glabrata does a haploid yeast, nondimorphic., don’t production pseudohyphae at 37 °C. C. glabrata to be more in people with low immunity state, C. glabrata to be a very opportunistic of the both urinary, genital organs and the blood circulating through the body of a person, while C. parapsilosis is a diploid yeast, dimorphic and don’t formation true hype C. glabrata. That is particularly widespread in people living with HIV and the aged [1]. In most surveys conducted in the US and Europe, C. glabrata is the second most common Candida species leading to invasive fungal infections [2]. By comparison, in Asian - Pacific countries and Latin America, C. tropicalis and C. parapsilosis are the second and third most common Candida spp., respectively [3].
Recently, there is a decrease in frequency of C. albicans and an increase in C. tropicalis and C parapsilosis, while the frequency of C. glabrata and C. krusei has remained unchanged .C. glabrata is also prevalent in the elderly, and frequency of C. parapsilosis decreases with age [2].
Candida is naturally present in humans and it does not cause any health problems in the host, but it works to destroy harmful bacteria. In individual who has good health, the proportion of Candida yeast is low due to the effectiveness of the immune system in addition to the presence of Probiotics bacteria such as Lactobacilli, however if the rate of these bacteria is low due to the defect of the immune system in the body or the use of antibiotic drugs, which leads to excessive growth of fungus, special Candida spp. and then change from the yeast to mycelium and then begin to penetrate the tissue host and causes candidiasis [4].
Yeast cells are approximately 2-10μm in the largest dimension, round to oval, and they are reproduced by budding. C. albicans is the lager yeast at 4-6 x 6-10μm, whereas C.glabrata and C. parapsilosis is the smallest at 1-4 x 2-9μm and 2-4 x 2-9μm, respectively [5].
Candida-associated with diarrhea in hospitalized patients, when there is a prior use of antibiotics. The yeast overgrowth is presumed to be antibiotics disruption of the normal flora. There is a mystery between Candida spp. Infection and the mechanism that causes diarrhea. It has been shown that Candida spp. can block lactase activity intestine that leads to lactose intolerance [6].
The Symptoms of Candida-associated diarrhea comprise lengthened diarrhea with abdominal ache, and suffer from colic but absence blood or mucus, nausea, vomiting, fever. The research and report showed that the clinical signs of the diarrhea were reduced and that coincides with the decrease fungus in the stool and the use of special fungicides [7].
Several molecular techniques have emerged for detection of Candida species which are based on amplification or hybridization of target nucleic acids, these are: Polymerase Chain Reaction (PCR) [8]. The genome of the fungi contain rRNA gene which is consisting of three subunits, these including small subunit (18S), 5.8S gene, and a large subunit (25S) gene, they are separated by the Internal Transcribed Spacer-1 (ITS1), and ITS2 regions . The ITS zone is a segment of non-functional RNA sequence which is located between rRNAs on a common precursor transcript, and it is usually used for identification of medically important fungi [8,9].
Material and Methods
Collection of samples
Hundred fecal specimens were collected from children under three years in cups. The patient’s name, age, gender were fixed in each cup and diagnosed immediately.
Culturing of samples
Sabouraud dextrose agar (SDA) was your utilized medium for the main isolation of Candida spp., it allows the growth of Candida and represses the expansion of many bacteria because of the low pH. Therefore; SDA was cooled down to 45-50 ºC then chloramphenicol antibiotic (250mg/liter) it was added then poured into sterile Petri dishes. SDA medium was inoculated with a small portion of the stool specimen with a sterile loop. The dishes remained incubated at 37 °C for 48hrs. And covered the plates by using the parafilm to preserve the culture and prevent contamination when saved in the refrigerator [10].
Chromogenic agar candida ( CAC)
Chrome agar Candida pure isolates of Candida were resuscitated by inoculating a loop full of culture from Sabouraud Dextrose Agar into 10ml sterile broth that incubated at 37 ºC for 3 days. After 72 hours of incubation, cultures were inoculated on the surface of the Chrome agar media by streaking a loop full of culture and incubated at 37 ºC for 72-96 hours with the plate facing upright. After 72-96 hours of incubation, all plates were removed from the incubator and the results were recorded. Candida colonies were initially identified by colonial color when compared with standard color photographs supplied by the manufacturer and also presented [11].
Molecular identification
DNA extraction: DNA was extracted from Candida spp. using Wizard Genomic DNA purification kit (Promega, USA) according to the protocol stated by the kit manufacturer.
DNA extraction procedure occurred by the following steps: Adding one ml of culture yeast for a period of twenty hours in the broth yeast extract-peptone-dextrose medium to a micro centrifuge tube. Centrifuging at 15,000 × g for two minutes to pellet the cells. Eliminate the supernatant. Resuspend the cells completely in 293μl of 50mM EDTA. Add 7.5μl of lysis buffer, then carefully mix it with a pipette. 5. Placing the specimen in the water bath at 37°C for one hour and cool to room temperature. Centrifuge at 15,000 × g for two minutes, after that, eliminate the supernatant. Adding Nuclei Lysis Solution amount 300μl to the cell pellet and then carefully mixing it with a pipette. Adding Protein Precipitation Solution amount 100μl and vortex quickly. Putting the specimen on the ice at five minutes. Centrifuging at 15,000 × g at three minutes. Preparing new microcentrifuge tube containing 300μl of the isopropanol then transfer supernatant containing DNA to it. 12. Inversing mixing continuously till the thread-similar strands of DNA from a noticeable piece. Centrifuging at 15,000× g for two minutes. 14. Emptying the supernatant and drain the micro centrifuge tube on new absorptive paper. Add 300μl of the 70% ethanol and invert the micro centrifuge tube sometimes to wash the DNA. Centrifuging at 15,000× g at two minutes and aspiration whole of the ethanol. 16. Draining the tube on clean absorbent paper and leave the pellet to air-dry for 12 minutes. Adding 50μl of DNA Rehydration Solution. 18. Adding 1.5μl of RNase Solution to the purified DNA sample. Vortex the sample for 1second. Centrifuge at 5 seconds to collect the liquid and incubate at 37°C for 15 minutes. Rehydrating the DNA by incubating the solution one night at 4°C. 20. Storing the DNA at 2–8°C.
Materials used for thermal cycling
Universal primers ITS1 (TCC GTA GGTGAA CCT GCG G) and ITS4 (TCC TCC GCT TATTGA TAT GC) were used to amplify the ITS1-5.8SITS2 fragment of fungal [12,13]. We’re synthesized by (Bioneer Co., USA) Optimization of PCR was accomplished after several trials. Thus the following mixture was adopted amplification reactions were produced in the 25μl final volume containing 12.5μl Go Taq® master mix (Promega, USA), 5μl of the primers and 5μl DNA template and complete the volume by Nuclease-Free Water. There are two programs thermal controller use in this study, Program for amplifying the ITS1-5.8S-ITS2 for Candida spp. to the identification of Candida spp. An initial denaturation step at 94 °C for five minutes was followed by twenty-five cycles of denaturation at 95°C for thirty seconds, annealing at 56 °C for fifty-four seconds , and extension at 72 °C for one minute, with a final extension step of 72 °C for Seven minutes [13] Program for amplify the 25S for Candida albicans to identification Candida albicans genotypes ,following thermal cycling: 94 °C for three minutes followed by thirty cycles of denaturation at 94 °C for one minute, annealing at 55 °C for one minute, extension at 72 °C for two and a half minutes and a 72 °C for ten minutes for a final extension following the last cycle [14].
Electrophoretic
Electrophoretic separation of DNA fragments was done according to the method described by Fankhauser [15]. Equipment: Ulra˗clean glassware, 1L beaker, 1L graduated; 250ml beakers cylinder, 100ml graduated cylinder horizontal, gel tray; electrophoresis unit, power supply, well comb, micropipettes, UV transilluminator. Supplies: latex gloves, 1X TBE buffer, agarose 1.5%, 5v/cm 2hr., micro centrifuge tubes [15].
Results
Cultural characteristics
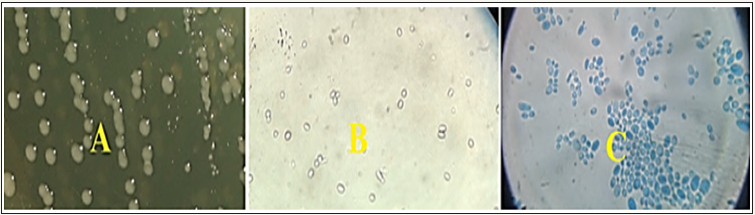
The results of specimens by the culture of SDA showed that 64 isolates of Candida infection. Morphological culture on SDA, colonies of Candida spp. colonies on sabouraud dextrose agar were white to creamy, round, soft, and smooth to wrinkled with a characteristic yeast odor, examination of Candida spp.isolates showed spherical to oval cells with a presence of budding and was larger than bacterial cells, as it appears in as is shown in (Figure 1).
Figure 1A: Candida spp. cultured on SDA at 37 ºC for 48 hrs,
B: Candida spp without stained (40X),
C: Candida spp stained with Lactophenol cotton blue (40X).
Chromogenic agar candida (CAC)
Figure 2: Colonies of Candida spp. cultured on chromogenic agar Candida at 37 ºC for 48 hrs, appeared easy distinction colors between C. albicans, C. krusei, C. tropicalis, but there are Colors are close to C. glabrata and C. parapsilosis.
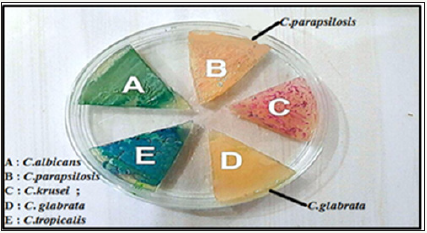
Where several colours emerged from the growth of Candida in that medium, were such as with the company protocol, greenish colonies of C. albicans, blue colonies of C. tropicalis, Purple- Pink colored colonies of C. krusei, C. glabrata produced cream to white and C. parapsilosis produced pinkish to white, as it appears in (Figure 2). In the present study, there were differences between Candida spp., 14/64 (21.9%) of the total isolates were identified Candida as C. glabrata, while 11/64(17.2%) isolate C. parapsilosis and 39/64 (60.9%) isolates were other Candida spp., C. albicans, C. krusei and C. tropicalis.
Molecular identification
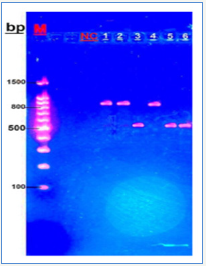
Fungus-specific universal primer pairs (ITS1 and ITS4) were able to successfully amplify the ITS1-5.8S-ITS2 region of all fungi tested, providing a single PCR product of approximately (510~870 bp) [13,16]. The 64 sample were molecularly identified according to ITS1-ITS4 sizes polymer chain reaction products for Candida spp. show that C. glabrata (871bp), while C. parapsilosis (520bp), as it appears in (Figure 3).
Figure 3: Agarose gel electrophoresis of Candida DNA products generated through primer ITS1 and ITS4, stained with ethidium bromide Lane M: Molecular marker (100bp); lane NC: negative control; lane, 2,4: C.glabrata; lanes 3,5,6: C.parapsilosis.
In this study, the results showed there are significantly (P< 0.05) relationship between C. glabrata and C. parapsilosis with children gender as shown in (Table 1). The results also showed there are insignificantly relationship between C. glabrata and C. parapsilosis with children and age group as shown in (Table 2). They also insignificantly relationship between glabrata and C. parapsilosis with children and mode of feeding as shown in (Table 3).
Table 1: Candida spp the infection rate among patients according to gender.
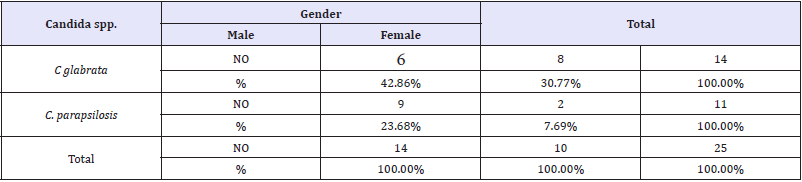
P-value= 0.0483 sig. (P<0.05)
Table 2: Candida spp. the infection rate among patients according to age.
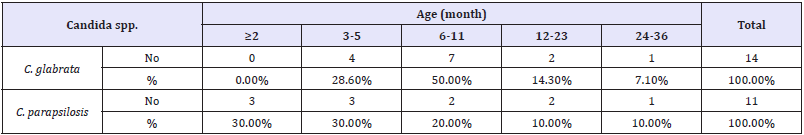
p-value =0.227 non sig. p>0.05.
Table 3: Candida spp. the infection rate among patients according to feeding.
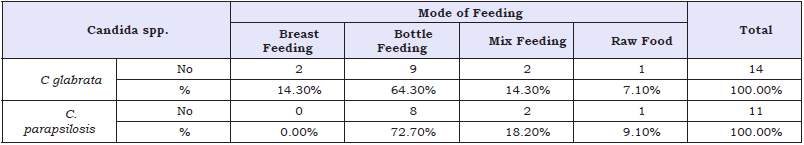
p-value= 0.631 non sig. (p>0.05)
Discussion
Intesar and Ahmed in Iraq, showed in their study from 23 isolates of Candida spp. were 3 3(9.4%) of both isolate of C. parapsilosis and C. glabrata that isolated from children with diarrhea [17]. A study by Zang L. et al. in China showed that C. glabrata was (20.3%) and C. tropicalis was (8.8%) [18]. In a study for identifying Candida spp. on chrome agar found 45 C. glabrata produced variously colored colonies on this medium, 29 White and 16 Lavender [19]. (2008) noted that 51 C. glabrata produced variously colored colonies on this medium, i.e., 37.8% maroon, 31.1% beige, 28.9% yellow, and 2.2% violet [20]. The ITS1 and ITS4 universal primer pairs were able to successfully amplify the ITS1-5.8S-ITS2 region of all fungi tested, providing a single PCR product of approximately (510~870bp) [13,16]. Our study agreed with Mirhendi e and Wahyuningsih , that found molecular identified according to ITS1-ITS4 sizes polymer chain reaction products for Candida spp. show that C. glabrata (871bp) and C. parapsilosis (520bp) [21].
Acknowledgment
Great appreciations are due to Research, College of Medicine - University of Diyala. I also thank the staff of laboratory and specialized doctors at AL-Batool Teaching Hospita, Baqubah, Iraq.
References
- Whelan WL, Simon S, Beneke ES, Rogers AL (1984) Auxotrophic variants of Torulopsis glabrata. FEMS microbiology letters 24(1): 1-4.
- Guinea J (2014) Global trends in the distribution of Candida species causing candidemia. Clin Microbiol Infect (6 Supp): 5-10.
- Reboli AC, Rotstein C, Pappas PG, Chapman SW, Kett DH, et al. (2007) Anidulafungin versus fluconazole for invasive candidiasis. N Engl J Med 356(24): 2472-2482.
- Ferreira C, Silva S, Faria-Oliveira F, Pinho E, Henriques M, et al. (2010) Candida albicans virulence and drug-resistance requires the O-acyltransferase Gup1p. BMC microbiol 10(1): 238.
- Remington JS (2011) Infectious diseases of the fetus and newborn infant. Saunders/Elsevier, USA.
- Gupta TP, Ehrinpreis MN (1990) Candida-associated diarrhea in hospitalized patients. Gastroenterology 98(3): 780-785.
- Levine J, Dykoski RK, Janoff EN (1995) Candida-associated diarrhea: a syndrome in search of credibility. Clinical infectious diseases 21(4): 881- 886.
- Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, et al. (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci 109(16): 6241-6246.
- Schwarzmüller T, Ma B, Hiller E, Istel F, Tscherner M, et al. (2014) Systematic phenotyping of a large-scale Candida glabrata deletion collection reveals novel antifungal tolerance genes. PLoS pathogens 10(6): e1004211.
- Deorukhkar SC, Saini S (2014) Laboratory approach for diagnosis of candidiasis through ages. International Journal of Current Microbiology and Applied Sciences 3(1): 206-218.
- Zarei MA, Drucker B, Mandall N, O’Brien K, Theaker E (2000) Isolation and identification of Candida species from the oral cavity using CHROMagar Candida. IBJ 2000 4(2): 57-61.
- White TJ, Bruns T, Lee SJ, Taylor JL (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 18(1): 315-322.
- Mirhendi H, Makimura K, Khoramizadeh M, Yamaguchi H (2006) A oneenzyme PCR-RFLP assay for identification of six medically important Candida species. Nihon Ishinkin Gakkai Zasshi 47(3): 225-229.
- Kaab HT (2014) Genotyping of Candida albicans Isolated from broilers by 25S rDNA Analysis. Kufa Journal for Veterinary Medical Sciences 4(1): 105-110.
- Zhong W, Shou Y, Yoshida TM, Babetta L (2007) Marrone Differentiation of Bacillus anthracis, B. cereus, and B. thuringiensis by using pulsed-field gel electrophoresis. Applied and environmental microbiology 73(10): 3446-3449.
- Ellis D, Davis S, Alexiou H, Handke R, Bartley R (2007) Descriptions of medical fungi. Nexus Print Solutions, Adelaide, South Australia, Australia.
- Juma IM, Mhawesh MA (2014) In vitro, Study of The Effect of Four Plant Aqueous Extracts on The Growth of Some Candida Species recovered from the stool samples of infants. Kufa Journal for Nursing Sciences 2(3).
- Zhang L, Zhou S, Pan A, Li J, Liu B (2015) Surveillance of antifungal susceptibilities in clinical isolates of Candida species at 36 hospitals in China from 2009 to 2013. Int J Infect Dis 33: 1-4.
- Ghelardi E, Pichierri G, Castagna B, Barnini S, Tavanti A, et al. (2008) Efficacy of Chromogenic Candida Agar for isolation and presumptive identification of pathogenic yeast species. Clin Microbiol Infect 14(2): 141-147.
- Ozcan K, Ilkit M, Ates A, Turac-Bicer A, Demirhindi H (2010) Performance of Chromogenic Candida agar and CHROMagar Candida in recovery and presumptive identification of monofungal and polyfungal vaginal isolates. Medical mycology 48(1): 29-34.
- Wahyuningsih R, Freisleben HJ, Sonntag HG, Schnitzler P (2000) Simple and rapid detection of Candida albicans DNA in serum by PCR for diagnosis of invasive candidiasis. J Clin Microbiol 38(8): 3016-3021.
© 2018 Sabah M Ali. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






