- Submissions

Full Text
Research & Development in Material Science
Exploring Common Domains in the Therapeutic Proteins with the Structure-Function Relationship Approach
Zahra Saeednejad and Soroush Sardari*
Drug Design and Bioinformatics Unit, Medical Biotechnology Department, Biotechnology Research Center, Iran
*Corresponding author:Soroush Sardari, Drug Design and Bioinformatics Unit, Medical Biotechnology Department, Biotechnology Research Center, Pasteur Institute of Iran, Tehran 13164, Iran
Submission: August 07, 2023;Published: September 08, 2023

ISSN: 2576-8840 Volume 19 Issue 3
Abstract
Proteins are composed of functional units and carry out their functions primarily through their constituent domains which are the independent folding groups. In this study, we explore the structure-function relationships among therapeutic proteins. We collected sequence data of the therapeutic proteins from Drug Bank and KEGG databases, then grouped different mechanistic and therapeutic classes. Each group was labeled based on structural functional groups in pharmaceutical applications by CATH database tool. This was performed by the domain information, functional families (Fun Fams), evolution, and the structural diversity of a superfamily, which is presented in CATH. Eight classes could be made for the various functionalities, and a corresponding correlation between each class and the relevant structure of the therapeutic protein category using RMSD scores was built. The most frequent functions were cytokine and protease activity that can be achieved by diverse folds of domains. These findings, not only help the structural biologist design better drugs but also clarify the structure-function relationship in those groups.
Introduction
Proteins have primary, secondary, and tertiary structures. Primary refers to the sequence of amino acids. Secondary proteins are local folded structures within a polypeptide due to the interactions between atoms of the backbone. The third and most remarkable level of proteins is the 3rd fold. Domains that can be considered unit structures have critical roles in protein function. Many key features of proteins can be found in these compact units. Domain activity relies on its three-dimensional structure or the third protein structure level.
Domains with globular structures are usually compact and by generalization for domains, these are also considered to fold individually, and form superfamilies with numerous similar members. It is evident that not all domains with a globular structure are the sole composing units that are functional for a protein. In addition, their detailed disorder formation can be of interest to research. By increasing protein data generation, it is shown that some similar protein groups may further present diversity among themselves. This variation may be to the extent that members of two distant groups could look close in some instances [1].
The possibility of persistent folds present in different domains can be observed in similar or even dissimilar proteins, structurally or functionally [2]. The binding site similarity may or may not relate to the similarity in the substrate. For example, out of 28 binding site categories, many have a variety of substrates, while in the category enzymes for binding to phosphoenolpyruvate, the conservation is very high. There are changes in size, along with variations in subgroups of domains. This could be the result of the loop or core structure changes [3].
Many computational studies are made to obtain structural information pertaining to the function of proteins. In building such structure-function correlations, the sequence data alone is not enough, and the scientists have to look into three-dimensional information as well. In these studies, a meaningful evolutionary relation can be obtained by computational study of the protein structures. Controversies over the structure-function relation exist among biologists but this claim is more accepted that most domains that share the same fold are associated with a single function [4]. The data based on the sequence and three-dimensional surface information are used to categorize proteins in various functional conservation sub-types [5].
The correlation between structure and function may not be represented by a one-to-one corresponding relation and there are always exceptions to indicate the depth of carry-over information inherited in structure or function layers are far more complex than previously thought. One such example is shown for the hydrolysis of glycosyl function that is instituted in various structures [6].
The algorithms to study domains can be based on sequence only, such as Pfam, or by considering deeper levels of structural information by looking into SCOP and CATH. Therefore, the details of classification can become more elaborate according to the level and volume of information considered in each database [1].
While it is usually true to have a particular structure handy in order to have a particular function in mind, as is the case for many mechanical tools made by humans, it is very easy to imagine the ability of various tools to be able to complete a particular task as our creativity or necessity would impose. Similarly, there is a many-to-many correlation in the protein structure-function world in place rather than a one-to-one stigma. This, however, would not limit our efforts to study and distinguish patterns in this field [7]. The reported normalized RMSD score showed to perform best in recognizing domains with the same fold [6,8].
Other early step efforts to establish the role of structure is to simply group the proteins based on their function and study the structures in each type [9]. The applications for such studies include the drug design process as well as basic research in cell and molecular biology [10].
The lack of a clear understanding of correlating structure and function would also limit our ability in drug development and building prediction models [7]. In our work, the structure-function relationship was studied with the means of domain comparison in the therapeutic protein which gave perspective regarding similar domains and RMSD range in the various groups.
Materials and Methods
In order to carry out domain analysis in therapeutic proteins, sequences were extracted from the Drug Bank, KEGG database, and the literature. Every domain in the FASTA format was matched to its corresponding InterPro and PDB code entry. Then using Swiss-Pdb Viewer due to the scope of domain comparison from a structural perspective. To further compare the sequence of proteins, we used PDB and SWISS-PROT tools [11].
Finding homologous domains
In order to investigate protein classifications, we used CATH. For this analysis, we used identical domains from the Structural Neighborhood of Representative and for a more accurate comparison, domains with the same RMSD were ignored. Comparison of the structure-function relationship to perform a study on the similarity between function (like those indicated by Fun Fam in CATH) and structure, similar superfamilies of therapeutic proteins with two or more proteins except for monoclonal antibodies (MAB) were considered.
While the domain itself is the basis for classification in CATH, the protein complex role, in general, is considered in Fun Fam. Therefore, in this study RMSD was the means of structure comparison and the whole protein for function comparison.
Results
Growth hormones
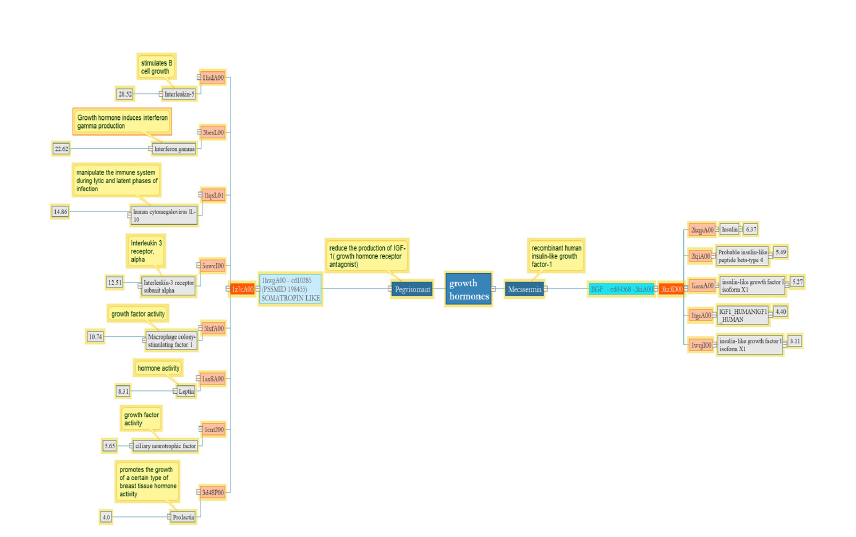
Pegvisomant as a growth hormone receptor antagonist is similar to Interleukin 5 and 10, Interferon-gamma, Interleukin receptor 3, Macrophage, leptin, and Prolactin in terms of threedimensional structure.
Cytokine binding is a similar feature among growth hormone receptor antagonists, interferons, and interleukins based on the UniProt database. While prolactin and Pegvisomant both regulate the receptor signaling pathway via JAK-STAT with about 4Å RMSD which is the lowest number in the group, this drug, and leptin with RMSD 8 have similar functions when binding peptides. It is apparent that in the group of Mecasermin or IGF-1, a structurefunction relation exists with RMSD 3-6.37. Therefore, domains in this group have a specific structure that can play a unique role in the biological mechanisms (Figure 1).
Figure 1:Pegisomant and Mecasermin are crucial growth hormones, each comprised of a single domain. Pegisomant has a wider range of RMSD, between 4-28, despite having only one domain, while the latter has five homologous domains with an RMSD range of 3-6.
Complement factor inhibitor
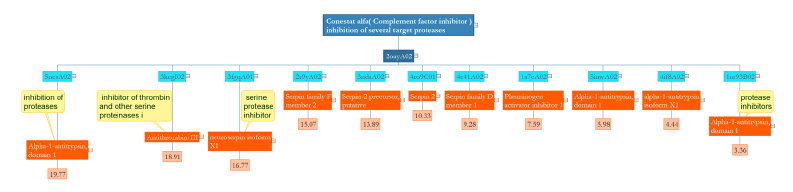
Conestat alpha or a recombinant human C1 esterase inhibitor with a single domain and average RMSD of 3-19 is similar to the serine protease inhibitor family and antitrypsin isoforms (Figure 2).
Figure 2:The main domain of Conestat alpha shares similarities with domains that possess protease activity. Protease activity can be attained with the lowest or highest RMSD.
Recombinant human deoxyribonuclease I
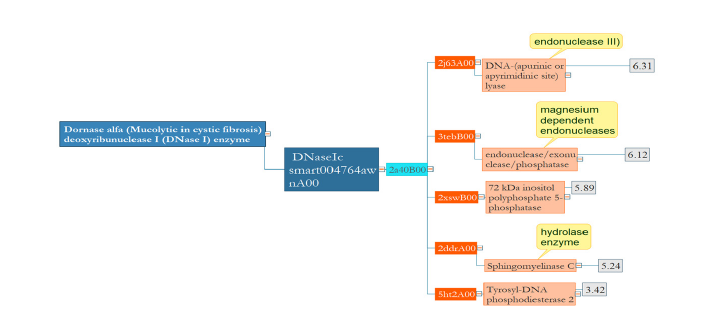
Dornase alpha which is used to break down extracellular DNA in the lungs has hydrolase activity like Tyrosyl-DNA phosphodiesterase 2 and Sphingomyelinase C. This single-domain protein seems to be conserved in terms of structure with the limited range of RMSD, 3-6 (Figure 3).
Figure 3:In Dornase alpha, there is no significant diversity between its homologous domains in terms of structure. Domains with the same structure do the same function.
Human interleukin 1 receptor antagonist protein
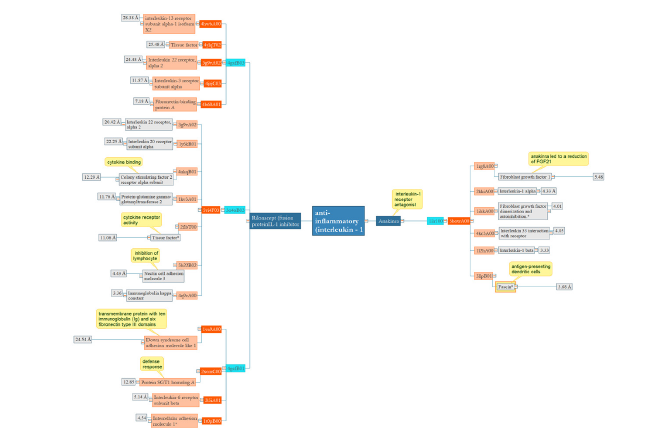
The therapeutic applications of human interleukin 1 receptor antagonist protein Anakinra include neonatal-onset multisystem inflammatory disease and rheumatoid arthritis. In this small group, the proteins are similar and have RMSD between 3.5-5. In addition, there is a clear relation between fibroblast growth and anakinra despite having the highest RMSD [12]. Rilonacept with three domains is used in the cryopyrin-associated periodic syndrome as an IL-1 blocker. First domain analysis: As in endothelial, induction of tissue factors occurs by IL-33, the result in the graph may be justified which shows the similarity between inflammation and coagulation factors with RMSD range 7- 28. Second domain analysis: This domain with a broad range of RMSD about 3-21 has the highest structural similarity to Nectin cell adhesion molecule and immunoglobin kappa constat. Third domain: There is no clear structural and functional similarity among this group with RMSD 4.5- 24.5 (Figure 4)..
Figure 4:Anakinra and Rilonacept are therapeutic proteins that participate in the anti-inflammatory system. While Anakinra is a single-domain protein, its domain shares both functional and structural similarities with five homologous domains.
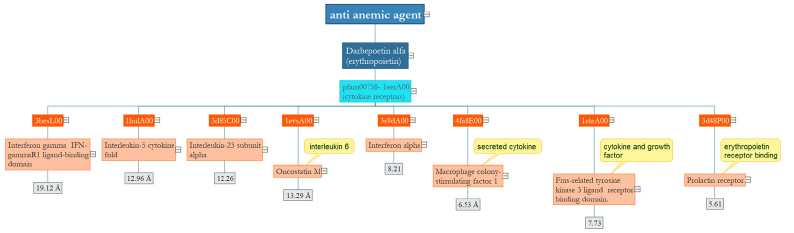
Ant-anemicagent
Darbepoetin alfa with a single domain stimulates erythropoiesis for the treatment of anemia and interleukins, interferon receptors, and prolactin receptor have common domains which are similar from a functional perspective. Darbepoetin decreases cytokine IL-6 and by affecting IL-10 can be with effects on patients with anemia and heart conditions [13]; (Figure 5).
Figure 5:The main domain of Darbepoetin alpha plays a role in cytokine activity. Mainly, similar domains with low RMSD have a similar function in the anemic agent.
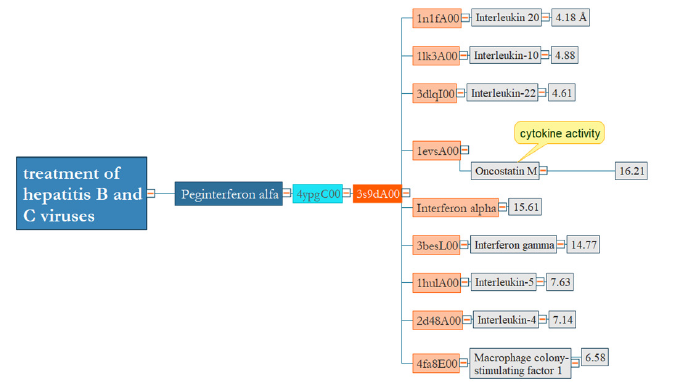
Treatment of hepatitis B and C viruses
Peginterferon alfa-2a is a modified form of human interferon with a single domain that is applied to induce the antiviral comeback for reducing hepatitis B and C conditions. Like the previous result, there is a strong relation among interferons and interleukins with RMSD about 4- 16 (Figure 6).
Figure 6:There are similar domains from a structural perspective between interferon and interleukins. However, peginterferon and interferon-gamma are not alike in terms of structure.
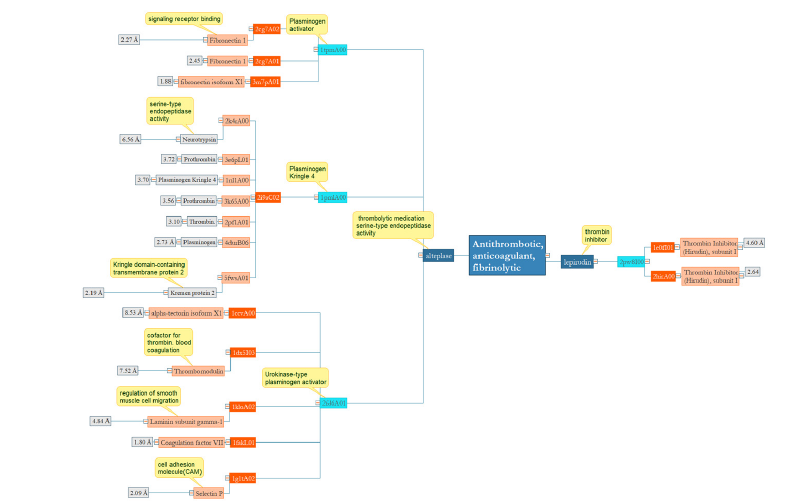
Antithrombotic, anticoagulant, fibrinolytic
Alteplase, a multidomain protein connects to fibrin-containing clots by the fibronectin finger-like moiety and the Kringle 2 part and is used in the urgent handling of heart muscle infarction, low oxygen stroke, and lung emboli. The first domain which acts as a plasminogen activator is like fibronectin with RMSD 1.8- 2.2 and is playing a role in the deprivation of the extracellular environment. Previous results indicate that fibronectin can be destroyed by urokinase in the absence of plasminogen. Plasminogen Kringle another domain of Alteplase is similar to prothrombin, thrombin, neurotrypsin, and Kremen protein with RMSD 2- 3.7 which indicates a structure-function relationship in this group.
The kringle group has a broad range of domain associates. These proteins carry out a range of purposes and can play as proteases (such as prothrombin or plasminogen,), protease initiators (tissue plasminogen activator or urokinase,), growth factors (hepatocyte and macrophage-inducing protein), and lipid carrier (apolipoprotein A). The last domain, the Urokinase-type plasminogen activator is highly similar to a P- selectin antagonist, as a helper to thrombolysis in acute heart muscle infarction with RMSD 2.09. Lepirudin, recombinant hirudin that acts as a highly specific thrombin inhibitor has a conserved domain that can be found only in similar structures (Figure 7).
Figure 7:Lepirudin acts as a thrombin inhibitor with one conserved domain in terms of structure and function. In alteplase, except third domain 2fdA01 as a Urokinase-type plasminogen activator, others have structure-function relationship.
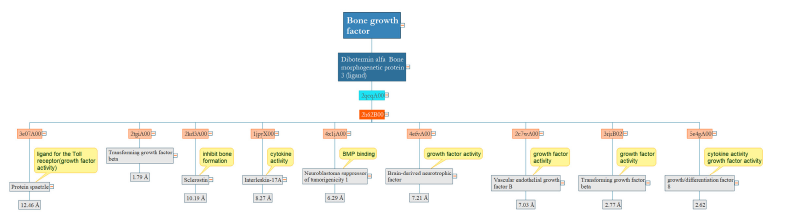
Bone growth factor
Dibotermin alfa with a single domain promotes bone and cartilage formation through anabolic effects in human osteoblastic cells. The similarity between this protein and Sclerostin, a neuroblastoma suppressor of tumorigenicity 1 in terms of structure and function is clear. However, the most similar domain from a structural perspective is the differentiation factor with RMSD 2.6 [14]; (Figure 8).
Figure 8:In terms of growth factor activity, Dibotermin alfa containing a single domain demonstrates similarity to homologous domains within a range of 2-7 RMSD. However, some domains with a greater RMSD range have a functional similarity.
Discussion
In this work, we focused on the structure-function relationship of biological drugs, which can be practical for structural biologists. Given the complexity of Considering the immense association between sequence, structure, and function in proteins, a practical approach is to investigate these connections in the background of separate groups and folds [9].
Therefore, we analyzed function conservation by using the structure of domains as core structures in the therapeutic proteins. In addition, we subsequently used RMSD in CATH database to compare the domain’s structures of homologous families. Strikingly, despite the examination of Todd et al., which was based on the relation of sequence identity and matching full (third EC level), we observe both enzymes and non-enzyme drugs in terms of the structure of the domains. Our results suggest that when clustering therapeutic proteins into similar families, distinction in function between homologs close groups may occur as Sangar found that, among similar proteins, the amount of different functions reduces intensely after a threshold of similarity in sequence at around 50% amino acid identity [15].
However, some protein functions are sufficiently general such that they can be achieved by different domain Structures [16]. our result shows that in growth factors, recombinant activated protein C and anti-anemic agent homologous families different domain folds have a general function such as cytokine, and protease activity. We use domain-based comparison since, in most therapeutic groups, the domain and the whole protein are corresponding entities. In addition, due to the shorter length of amino acids, the result of the comparison would be more accurate. In fact, based on previous research the application of thresholds for domains strikingly surges protein coverage due to the advanced quantity of usable pairwise associations. Furthermore, far relations can deliberate functional annotation in similar domains with equal structures in proteins with many domains [15]. There are some controversial results when comparing structure-function relation in the therapeutic proteins like lower RMSD numbers and higher ones do not correlate with function similarity. However, some proteins with a limited range of 2-10 A like Mecasermin, Dornase alpha, and Anakinra, the second domain of Alteplase, have similarities in terms of function. In contrast, others with a wide range of about 3-50 like Dibotermin alpha, peginterferon, darbepoetin, rilonacept, Conestat alpha, and Drotrocogin alpha. Pegvisomant despite having low RMSD, functions are not similar.
A fold association between proteins stands up in two methods: the proteins contain a shared ancestor [8] or as affiliates of the identical structural group [10]. or the proteins have united to the same group without a shared evolutionary past [17].
The advantage of the superfamily-specific - based method at the domain level over the use of general thresholds is based on the noticeable, non-basic association between structure and function (function conservation). The functional differences can be detected at the low number of RMSD, whereas functionally comparable proteins can have a high number of likenesses. This is due to dissimilar evolutionary paste for various proteins and domain super-families (similar); furthermore, the diverse functions of the super-fold structures suggest that they may have arisen independently and not by divergent evolution [18]. For example, although Alteplase as a thrombolytic medication is similar to Selectin P in terms of structure with 2.09A, there is no functional similarity. Since CATH provides all-inclusive structural and functional tagging for sequences from the main protein data banks such as Ensembl and UniProt [19].
we also generated data based on the categories of therapeutic proteins which help us to have a better inside regarding their pharmacological actions. Considering these classification factors, the thresholds might turn out to be particularly valuable for the attributing of uncharacterized sequences to precise pathways and play a role as an extra assurance measure in the reconstruction of pathways [15]. Understanding biological sequence-structurefunction relationships and finding core domains in therapeutic proteins give biologists a unique look at the production of therapeutics. Once a 3D structure of therapeutic protein gets assigned domains, for example, automatically by CATH source, the scientists can predict functional characters for a structure according to the similar RMSD range. This approach in medicine can lead to a better drug design by using CATH more efficiently which is the result of having better insight into common domains.
References
- Lees JG, Dawson NL, Sillitoe I, Orengo CA (2016) Functional innovation from changes in protein domains and their combinations. Curr Opin Struct Biol 38: 44-52.
- Holm L, Sander C (1998) Dictionary of recurrent domains in protein structures. Proteins 33: 88-96.
- Todd AE, Orengo CA, Thornton JM (2001) Evolution of function in protein superfamilies, from a structural perspective. J Mol Biol 307(4): 1113-1143.
- Lee D, Redfern O, Orengo C (2007) Predicting protein function from sequence and structure. Nature Reviews Molecular Cell Biology 8: 995-1005.
- Orengo CA, Todd AE, Thornton JM (1999) From protein structure to function. Curr Opin Struct Biol 9(3): 374-382.
- Gherardini PF, Helmer-Citterich M (2008) Structure-based function prediction: approaches and applications. Brief Funct Genomic Proteomic 7: 291-302.
- Sadowski MI, Jones DT (2009) The sequence-structure relationship and protein function prediction. Curr Opin Struct Biol 19(3): 357-362.
- Redfern OC, Harrison A, Dallman T, Pearl FMG, Orengo CA (2007) Cathedral: A fast and effective algorithm to predict folds and domain boundaries from multidomain protein structures. PLOS Computational Biology 3(11): e232.
- Redfern OC, Dessailly B, Orengo CA (2008) Exploring the structure and function paradigm. Curr Opin Struct Biol 18(3): 394-402.
- Reis R, Moraes I (2019) Structural biology and structure-function relationships of membrane proteins. Biochem Soc Trans 47(1): 47-61.
- Pearson WR, Lipman DJ (1988) Improved tools for biological sequence comparison. Proceedings of the National Academy of Sciences of the United States of America 85(8): 2444-2448.
- Ebrahimi F, Urwyler S, Betz M, Christ E, Schuetz P, et al. (2019) Effects of anti-inflammatory treatment on fibroblast growth factor-21 in obesity and metabolic syndrome. Endocrine Abstracts.
- Kourea K, John TP, Dimitrios F, Fotios P, Ioannis P, et al. (2008) Effects of darbepoetin-alpha on plasma pro-inflammatory cytokines, anti-inflammatory cytokine interleukin-10 and soluble Fas/Fas ligand system in anemic patients with chronic heart failure. Atherosclerosis 199 (1): 215-221.
- Nakayama S, Taiji Y, Nobuya H, Toshikazu A, Uta N, et al. (2017) Transforming growth factor β- and interleukin 13-producing mast cells are associated with fibrosis in bone marrow. Human Pathology 62: 180-186.
- Addou S, Rentzsch R, Lee D, Orengo CA (2009) Domain-based and family-specific sequence identity thresholds increase the levels of reliable protein function transfer. J Mol Biol 387(2): 416-430.
- Leman JK, Szczerbiak P, Renfrew PD, Gligorijevic V, Berenberg D, et al. (2022) Sequence-structure-function relationships in the microbial protein universe. Nat Commun 14(1): 2351.
- Moult J, Melamud E (2000) From fold to function. Current Opinion in Structural Biology 10(3): 384-389.
- Orengo CA, Jones DT, Thornton JM (1994) Protein superfamilies and domain super folds. Nature 372: 631-634.
- Sillitoe I, Bordin N, Dawson N, Waman VP, Ashford P, et al. (2020) CATH: increased structural coverage of functional space. Nucleic Acids Research 49(D1): D266-D273.
© 2023 Soroush Sardari. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






