- Submissions

Full Text
Perceptions in Reproductive Medicine
Human X-linked CUL4B Gene Mutation Mediated Changes in Function during Development of Testis after Prediction of 3D Structure
Ajit Kumar Saxena1*, Agarwal M2, Ramanuj K1, Aniket K1 and Singh CK1
1Department of Pathology/Laboratory Medicine, India
2Department of Obstetrics/Gynecology, India
*Corresponding author: Ajit Kumar Saxena, Department of Pathology/ Laboratory Medicine, India
Submission: September 07, 2020;Published: September 28, 2020

ISSN: 2640-9666Volume4 Issue1
Abstract
Introduction: CUL4B gene is ubiquitin ligase belongs to the Cullinring ubiquitin ligase family. In recent study we have identified a series of MTNR1B, SENP3, AKAP3 and PLOD3 genes and their functional interaction to the ligand binding capacity after prediction of 3D structure to the drug like methotrexate, which plays a significant role during spermatogenesis. Although, the role of Cul4B gene mutation and its impact to the ligand binding during development and differentiation of male gonad (testis) remains unexplored in fertility.
Objective: The present study has been designed to characterize the mutation of CUL4B gene sequence and correlate with functional aspect to the ligand binding site to understand the translational event of non-frame shift mutated (deleted) nucleotide sequence GGAGGA of DNA. The findings of mutations try to correlate with anatomical feature (testicular size) of the gonad and fertility.
Material and methods: Blood samples were collected from the cases of clinically diagnosed non obstructive azoospermia (NOA) with respective age matched controls. Study was carried out using whole genome sequencing (NGS) from Illumina (USA) to characterize the nature of mutation. The study was further extended to develop 3D structural analysis of the candidate gene CUL4B and protein (ligand) binding activity with drug like methotrexate using molecular docking (iTASSER) techniques for gene coded functional changes in testis.
Results: Findings of NGS DNA sequencing confirm the non - frame shift mutation in homozygous condition of CLU4B gene involving deletion at position 1761-1746 of GGAGGA nucleotide sequences. After decoding of mutation region (gene) confirm loss of non-essential amino acid glycine glycine resulting changes in physiological function during spermatogenesis in testis. The predicted 3D helical structure of CLU4B gene showing binding of amino acid residue to ligand (methotrexate) as model to evaluate the functional activity of the gene by using bioinformatics tools.
Conclusion: DNA sequencing analysis confirms the non-frame shift mutation of CUL4B gene in non-obstructive azoospermia male in homozygous condition. After mutation of GGAGGA sequence confirm either loss or deletion of non-essential amino acid residue (glycine-glycine) failed to bind to the ligand respective ligand (protein) resulting interference to the development of testis and infertility.
Keywords: CUL4B gene; Infertility; DNA sequencing; 3D protein structure
Introduction
Human infertility is a highly complex phenomenon and etiological factors are poorly understood [1]. CUL4B gene is a member of the family of Cullin scaffold proteins which makes a series of ubiquitin-protein ligase complexes that regulate the degradation of cellular proteins [2]. The CUL4B gene is mapped on Xq24, is and consist of 22 exons which encodes a protein of 913 amino acids (https://www.ncbi.nlm.nih.gov/gene/?term=cul4b). The cullin domain is located between amino acid residues 14 and 354 and is characterized by a C-terminal globular domain (cullin homology domain). Earlier study of CUL4B gene point mutation assigned on X- chromosome in homozygous condition of has been associated to male infertility [3]. CUL4B gene hypothesized to play an important role in maintaining structural and functional role during spermatogenesis. CUL4 gene family have two members, CUL4A and CUL4B play a crucial role for the survival of both male and female germ cells.
CUL4B is expressed in testis during spermatogenesis and mostly in post-meiotic spermatids stage and hardly expressed in spermatocytes [4,5]. CUL4B gene expression has distinct functions during development of gonad, but still to date, the crucial function(s) of CUL4B gene has not been clear in association with infertility. The rationale behind this study to recognize the impact of non-frame shift gene mutation of CUL4B and their interaction to ligand (protein) binding after prediction of 3D structure using known MTX drug as a model of infertility using molecular docking techniques. The present study might be helpful to explore the mechanism of abnormal differentiation of male gonad (testis) after gene protein drug interaction if mother exposed antenatally with anticancer molecules which might have interfare to the spectrum of cellular events of spermatogenesis and fertility.
Material and Methods
In the present study, clinically diagnosed cases of male infertility referred from OPD of AIIMS, Patna (Bihar, India) for Genetic investigations. The study was approved by Institutional Ethical Committee (IEC) and blood samples were collected by written informed consent from guardian. These cases of infertility are mainly categorized in three different groups i.e. non-obstructive azoospermia, obstructive azoospermia and oligozoospermia on the basis semen analysis by WHO [6] where the sperm count >20×106/ml, progressive motility>50% and normal morphology >30% and established fertility (with one or more children) were included as controls. All patients were initially evaluated by clinician and conventional medical diagnostic including patient’s background and physical examination analyses were performed. None of them had any history of childhood disease, environmental exposure, radiation exposure or prescription drug usage that could account for their infertility. The median age of patients included in the study was 35.4 years.
Isolation and characterization of mutation using whole exome sequencing (WGS)
Genomic DNA was isolated from clinically diagnosed cases of non-obstructive azoospermia and blood sample (1.0 ml) were collected after written informed consent and according to procedure of DNA isolation kit (Promega USA). Quantitative analysis of the DNA (ng/ul) was measured by Nanodrop spectrophotometer (Thermo. USA) and mutations of microdeletion of Y- chromosome regions (AZF a, b, c) were confirmed using PCR procedure before performed the sample for sequencing. After confirmation of mutation, genomic DNA was purified before initiation of sequencing, performed by Illumina HiSeq 2000 platform performed by Xcelris Lab at Ahmadabad, India. The sequencing data was analysed through http://www.ensembl.org/Homo_sapiens/Gene and mutated region identifies by http://www.ensembl.org /Homo_sapiens/Tools/Blast/Alignment. During data analyses of candidate gene variants when compared to genome-wide data using external available databases such as the Uniprot (https://www.uniprot.org/uniprot/Q13620). The data was further analysed using large-scale exome sequencing including ensemble database for structural and functional (http://www.rcsb.org/structure/4A64) effects of the mutated region by pymol visualizing software [7].
Homology modelling by I-TASSER
The most common server is a hierarchical approach of using I-TASSER (Iterative Threading Assembly Refinement) based on algorithms for the prediction of protein structure and function. It first identifies structural templates from the PDB by multiple threading approach LOMETS, with full-length atomic models constructed by iterative template-based fragment assembly simulations. Function insights of the target are then derived by re- threading the 3D models through protein function database BioLiP. Recently, I-TASSER server predict protein structure in community wide Critical Assessment of Techniques for Prediction (CASP) protein structure such as CASP7, CASP8, CASP9, CASP10, CASP11, CASP12, and CASP13 experiments [8]. The most common antagonist of folate is methotrexate (MTX), an antineoplastic and immunosuppressant drug prescribed by large number of clinicians. It acts as an inhibitor of zn enzyme the tetrahydrofolate dehydrogenase and prevents the formation of tetrahydrofolate form dihydrofolate in the presence of an enzyme reductase required for DNA synthesis. Molecular docking is an essential technique for prediction of protein and ligand interaction which help to determine accuracy of complex geometry based on free binding energy and charges using auto dock tools software. Auto Dock parameter help to arrange distance-dependent dielectric functions were used for the calculation of van der Waals and the electrostatic forces for stable binding [9]. The non-polar hydrogen atoms were merged and rotate during docking calculations with drug (MTX) ligand protein model.
Result
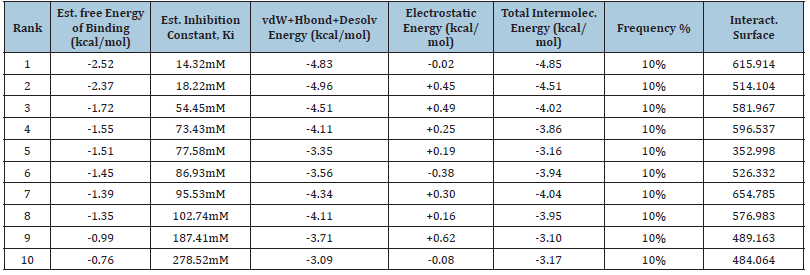
Cytogenetic location of chromosome-X (Homo sapiens annotation of cytogenetic GRCh38.p12) (https://www.ncbi.nlm.nih.gov/genome/tools/gdp) shown in Figure 1A, showing chromosomal loci of CUL4B gene mapped on Xq24 region. The DNA sequencing data reveals that CUL4B gene mutation occurs in homozygous condition with deletion of GGA GGA nucleotide sequences at position 1741-1744, which encodes a protein consist 913 amino acids, although, the total length of nucleotide consist of 51241bp is depicted in Figure 1B. The whole exome sequencing is a powerful technique for the identification of de-novo mutations. After validation and confirmation of CUL4B mutated regions in azoospermic cases of infertility the different biological databases NCBI (https://www.ncbi.nlm.nih.gov/protein/?term=Homo+sapiens+CUL4B), Ensembl (http://asia.ensembl.org/Homo_sapiens/Gene/Summary?db=core;g=ENSG00000158290;r=X: 120524609-120575794). This non frame shift mutation (deleted) region of DNA sequences after decoding failed to transcribed into non-essential amino acid - glycine glycine (two times) in the case of NOA as shown in Figure 1C. The prediction of 3D structure of CUL4B gene coded protein is documented in protein data bank (https://www.rcsb.org). Figure 2A & 2B showing the normal and mutated amino acid sequence based on NCBI data based (normal sequence) and NGS based DNA sequence (for mutated sequence) of CUL4B protein. Further protein sequence has been confirmed by using Uniprot (https://www.uniprot.org/blast/?about=Q13620[1-913] & key=Chain & id=PRO_0000393946). Figure 2C & 2D showing normal and mutated helical structure of CUL4B protein obtained from protein data bank highlighting normal and mutated (loss of glycine) amino acid sequence. Therefore, molecular docking was performed to evaluate the interaction with CUL4B encoded normal and mutated protein to methotrexate as shown in Figure 3A & 3B. Further, docking calculations were performed using Gasteiger charges method for the study of free binding energy based on normal and truncated protein (mutated protein) interaction with drug (methotrexate) and the findings are depicted in Table 1A & 1B. Table-1B showing the accuracy of docking findings in the mutated protein i.e. -2.52kcal/mol. considering the most populated and lowest energy out of ten different binding sites that were predicted, while normal protein interaction with MTX showed higher free binding energy (-1.33kcal/mol) as compared to the mutated protein (Table 1A). Thus, free binding energies suggested a better interaction between drug (MTX) with the mutated CLU4B protein as compared to the normal structure.
Figure 1: Schematics representation of chromosome loci of CUL4B gene on Xq24 (figure 1A), details of sequencing of CUL4B showing non frame shift gene mutation (figure 1B) and after decoding the nucleotide sequence GGAGGA in to corresponding amino acid (glycine) as shown in figure 1C.
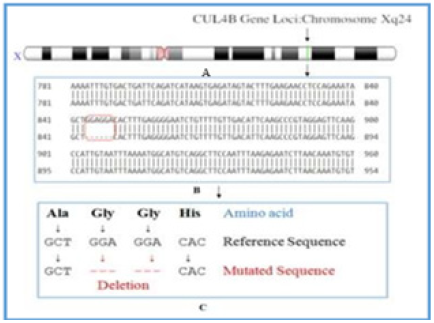
Figure 2: Showing CUL4B gene coded amino acid sequences normal (figure 2A) and mutated (figure 2B) and their 3D helical protein structure between normal (figure 2C) and mutated region as showing loss of two molecule of glycine as shown in figure 2D.

Figure 3: Showing normal CUL4B 3D protein structure with binding pocket to the amino acid residues (blue) and methotrexate (MTX) during ligand binding molecule (green ball & stick) having best interaction hydrogen bonding (red) in normal (figure 3A) and mutated structure (figure 3B).

Table 1A: Illustrate the CUL4B protein free binding energy with MTX ligand molecule with ten different binding energies. (A) Binding energies ranging from -1.33kcal/mol to -0.12kcal/mol in the case of normal CUL4B with MTX. (B) Binding energies ranging from -2.52kcal.mol to 0.76kcal/mol in the cases of mutated CUL4B with MTX. The ranks are shown 1-10 on the basis of the best fit protein ligand structure in the case of normal protein.
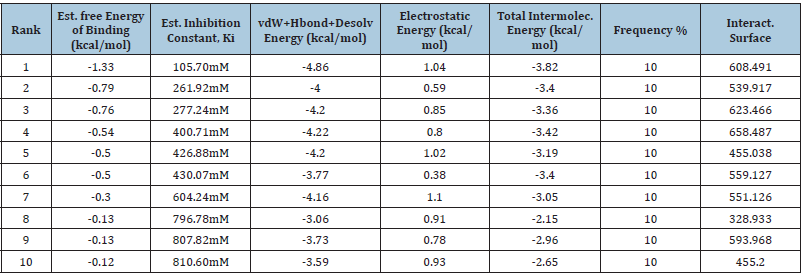
Table 1B:
Discussion
In humans genetic and epigenetic factors are involved in the differentiation and development of male gonads, while secondary sexual characters are based on endocrine regulation (testis). Present study explore the association of the E3 ubiquitin ligase CUL4B gene and deletion (loss) of GGA nucleotide sequences followed by change of translation event i.e. loss of non-essential amino acid (glycine) play an essential role during spermatogenesis. Genetic factors (mutation) modulate the events of germ cell proliferation and further differentiation in Testis. Despite of the major role of genetic factors contributing male infertility still remains a major challenge for the clinicians as well as the scientist in substantial proportion of cases. At the same time noncoding regions of the genome should not be forgotten in the biomedical research and consequently may be helpful to identify the unknown genetic factors in the same cases of Infertility. Interestingly, the non-coding regions were found to be useful for clinical diagnosis of unexplained cause of male infertility [10]. Furthermore, environmental factors including arsenic exposure are also responsible for reproductive dysfunction affecting male fertility [11]. During spermatogenesis, CUL4B is expressed in spermatogonia, differentiating secondary spermatocytes and spermatids during recombination and crossing over events. In human CUL4B deficiency causes development of hypogonadism in males [12]. Earlier, studies by our group identified the involvement of several genes regulating spermatogenesis including X linked ARSD (Arylsulfatase D) gene polymorphism [13-16]. In addition to these, several genetic polymorphisms have been demonstrated to be significantly associated risk factor with male infertility [17]. By the end of 19th century new technology emerged like NGS data can also be used to identify new mutations and their linkage associated homozygosity in copy number variations at specific intervals in various disease like infertility [13,18,19]. Recent advancement in molecular biology techniques including whole exome sequencing (WES) has emerged as a most reliable tools for the identification of “new gene mutations” based on SNP, insertions/deletions (indel mutations). It is also a cost-effective tool for detecting rare mutation within a patient population. More than 30 Mendelian disease genes have been identified based on NGS, including recessive and dominant character in the proband.
Due to upregulation of CUL4B gene coded protein and their associated functional aspects in the cases of spermatogenesis remain obscure. Present study identified the predicted 3D structure of CUL4B and their functional aspect in the case of NOA. Recently, a lot of techniques are available for the study of structural and functional analysis of biomolecules. Each molecule has their own specific structure and function based on capability to the ligand binding active sites. The mutational analysis of CUL4B truncated protein with ligand binding like MTX showing more significant free binding energy (-2.52kcal/mol) as compare to normal (-1.33kcal/mol). The lowest energy confirmed of each molecule which lying 0 to -10kcal/mol were subjected to standard docking calculations for ligand binding [20]. The CUL4B gene encoded protein provide valuable knowledge to understand the molecular dynamic of their structural and functional role in spermatogenesis during gene-protein-drug (MTX) interaction. Methotrexate (MTX) is a highly toxic molecule and entered into S phase of cell-cycle to inhibit DNA synthesis followed by cell death. Present study also help to explain that after mutation of CUL4B gene might have lost their binding ability to the respective ligand (protein) resulting in altered developmental processes of male gonad (testis) and thus affecting germ cell population (spermatocytes or spermatids).
In the present study, genetic factors for male infertility become prime important to understand the interaction between candidate gene CUL4B and their translation event (loss of glycine). Thus, the molecular modelling help to explain the physiological function based on ligand bindings sites to the target proteins (mutated). Thus, authors conclude that, present study suggests that CUL4B gene mutation is responsible not only for the size of testis but may interfare to the events of spermatogenesis, if foetus exposed antenatally to teratogen like methotrexate. Hence, such molecules might have interfare to the differentiating male gonads (testis) results increase “risk” of male infertility.
Acknowledgement
AKS thankfully acknowledges to the Director, AIIMS Patna for valuable suggestions, and financial support is provided by the Department of Biotechnology (Govt. India). No.BT/PR14671/ MED/12/487/2010) to carry out this research work.
References
- Ferlin A, Arredi B, Foresta C (2006) Genetic causes of male infertility. Reprod Toxicol 22(2): 133-141.
- Tarpey PS, Raymond FL, Meara OS, Edkins S, Teague J, et al. (2007) Mutations in CUL4B, which encodes a ubiquitin E3 ligase subunit, cause an X-linked mental retardation syndrome associated with aggressive outbursts, seizures, relative macrocephaly, central obesity, hypogonadism, pes cavus, and tremor. Am J Hum Genet 80(2): 345-352.
- Carrell DT, De Jonge C, Lamb DJ (2006) The genetics of male infertility: A field of study whose time is now. Arch Androl 52(4): 269-274.
- Kopanja D, Roy N, Stoyanova T, Hess RA, Bagchi S, et al. (2011) Cul4A is essential for spermatogenesis and male fertility. Dev Biol 352(2): 278-287.
- Boh BK, Ng MY, Leck YC, Shaw B, Long J, et al. (2011) Inhibition of cullin RING ligases by cycle inhibiting factor: Evidence for interference with Nedd8-induced conformational control. J Mol Biol 413(2): 430-437.
- Cooper TG, Noonan E, von Eckardstein S, Auger J, Baker HW, et al. (2010) World Health Organization reference values for human semen characteristics. Hum Reprod Update 16(3): 231-245.
- De Lano, WL, Pymol (2002) An open-source molecular graphics tool. CCP4 Newsletter on Protein Crystallography 40: 82-92.
- Yang J, Yan R, Roy A, Xu D, Poisson J, et al. (2015) The I-TASSER Suite: protein structure and function prediction. Nat Methods 12(1): 7-8.
- Bikadi Z, Hazai E (2009) Application of the PM6 semi-empirical method to modeling proteins enhances docking accuracy of AutoDock. J Cheminform 1: 15.
- Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS (2010) Non-coding RNAs: regulators of disease. J Pathol 220(2): 126-139.
- Saxena AK, Kumar A (2020) Fish analysis for drug and chemicals mediated cellular toxicity. Springer briefs in applied science and technology. In Chapter- 5. Effect of arsenic exposure in reproductive health pp. 59-80.
- Kerzendorfer C, Hart L, Colnaghi R, Carpenter G, Alcantara D, et al (2011) CUL4B-deficiency in humans: understanding the clinical consequences of impaired Cullin 4-RING E3 ubiquitin ligase function. Mech Ageing Dev 132(8-9): 366-373.
- Saxena AK, Tiwari M, Agarwal M (2018) Single nucleotide polymorphism of Arylsulfatase D gene (ARSD) and their association with male infertility. J Clin Gen Genomics 1(2): 11-13.
- Saxena AK, Tiwari M, Kumar A (2019) Penetrance of de novo mutation of USP9Y and PCDH11Y gene in AZF regions of non-obstructive Azoospermic population in India. Int J Curr Res 11(2): 1373-1379.
- Saxena AK, Aprajita, Kumar A, Tiwari M (2019) Impact of MTNR1B and SENP3 Gene on Male Infertility: Prediction of 3D Structural Modelling of Protein and Drug Interaction during Spermatogenesis. J Mol Genet 2(1): 1-8.
- Saxena AK, Tiwari M, Agarwal M, Aniket Kumar A (2020) Prediction of 3D Protein Structure based on the mutation of AKAP3 and PLOD3 Genes in the case of Non-Obstructive Azoospermia. Int J Fertility and Sterility 14(2): 102-109.
- Ceylan GG, Ceylan C (2015) Genetics and male infertility. World J Clinical Urology 4: 38-47.
- Krawitz PM, Schweiger MR, Rödelsperger C, Marcelis C, Kölsch U, et al. (2010) Identity-by-descent filtering of exome sequence data identifies PIGV mutations in hyperphosphatasia mental retardation syndrome. Nature Genetics 42: 827-829.
- Becker J, Semler O, Gilissen C, Li Y, Bolz HJ, et al. (2011) Exome sequencing identifies truncating mutations in human SERPINF1 in autosomal-recessive osteogenesis imperfecta. Am J Hum Genet 88(3): 362-371.
- Alvarez CA, Costa AM, Vilarrasa J (2017) The performance of several docking programs at reproducing protein-macrolide-like crystal structures. Molecules 22(1): 136.
© 2020 Ajit Kumar Saxena. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






