- Submissions

Full Text
Progress in Petrochemical Science
Microbial Diversity of Petroleum Polluted Soil at Ayetoro Community in Ilaje Riverine Oil Producing Areas of Ondo State, Nigeria
Ajayi AO* and Abiola AK
Department of Microbiology, Adekunle Ajasin University, Nigeria
*Corresponding author: Ajayi AO, Department of Microbiology, Adekunle Ajasin University, Akungba-Akoko, Ondo State, Nigeria
Submission: April 24, 2018;Published: May 14, 2018

ISSN 2637-8035Volume1 Issue5
Abstract
This study shows the potential of various types of bacterial isolates that utilizes hydrocarbon substrates which enhances the degradation of this pollutant in the environment. Soil samples used for this purpose were obtained from oil-producing Ayetoro community in Ilaje Local Government of Ondo State, Nigeria. Bacterial isolates were recovered by culturing them on nutrient agar and Mineral Salt Medium (MSM). Of the twenty two (22) bacterial isolates, thirteen (13) were Gram positive and nine (9) were Gram negative. The Gram positive bacteria were identified by Gram staining and biochemical tests while the Gram negative bacterial isolates were identified with the use of API® identification kit. Five (5) of thebacterial species including Micrococcus luteus, Bacillus cereus, Bacillus subtilis, Pseudomonas aeruginosa and Acinetobacterlwoffi exhibited potential to degrade Bonny light crude oil by showing visible growth around the oil in MSM using the hole-plate diffusion method. The study also showed microbial growth indicated by varying turbidity on MSM broth incorporated with Bonny light oil with optical density range of 0.18-0.85 at 540nm. From this study, Pseudomonas aeruginosa showed the highest optical density of 0.76-0.85 and has the greatest potential to degrade hydrocarbon. Thus, these oil-degrading properties of the microorganisms informed the choice of this group of organisms for bioremediation of oil-polluted sites.
Keywords: Bioremediation; Hydrocarbon; Microbial isolates; Nigeria; Oil pollution
Introduction
Petrochemical hydrocarbons are valuable source of energy around the world for our day to day activities and industrial developments. Nigerians use petrol as one of its world’s most important derived energy source [1]. Nevertheless, civilization and activities of petroleum and petroleum associated industries in Nigeria and other parts of the world had led to increase in oil pollution in our environment. Leaks and accidental spills occur regularly during the exploration, production, refining, transport and storage of petroleum and petroleum product. The amount of natural crude oil seepage was estimated to be 600000 metric tons per year with a range of uncertainty of 200000 metric tons per year [2]. In this context, pollution is defined as the addition to any segment of the environment, any material which has detrimental effect on the ecosystem [3].
Crude oil exploration causes great environmental problem in Nigeria as a result of oil spillage both onshore and offshore. It is known that greater degradation of oil pollutant is carried out insitu by a consortium of microorganism and more than 200 species of bacteria and fungi and even biodegrade hydrocarbons [4]. The various genera that have been reported to contain hydrocarbon degrading species include Pseudomonas spp, Vibrio spp, Corynebacterium spp, Arthrobacter spp, Brevibacterium spp, Flavobacterium spp, Sporobolomyces spp, Achromobacter spp, Bacillus spp, Aeromonas spp, Thiobacillus spp, Lactobacterspp, Staphylococcus spp, Penicillium spp and Articulosporiusm spp. These organisms has been isolated in large numbers from many oil polluted water and soil but are found in less number in uncontaminated environment [5].
The process of bioremediation is defined as the use of microorganisms to detoxify or remove pollutants [6]. In addition, bioremediation technology is believed to be non-invasive and relatively cost effective [7]. By nature, populations of microorganisms represent one of the primary mechanisms by which petroleum and other hydrocarbon pollutants can be removed from the environment and are cheaper than other technologies [8]. The principle by which a consortium of microorganisms acts to bring about the oxidation of complex compound is known as cometabolism. This principle is employed by oil companies in Nigeria to remediate oil polluted sites in a process known as remediation by enhanced natural attenuation (RENA). This is because the microbial degradative mechanism appears to be some natural processes which eliminate the bulk of oil pollutant after initial physical and chemical breakdown has occurred. The success of oil spill bioremediation depends on one’s ability to establish and maintain condition that favors enhanced oil biodegradation rate in the contaminated environment [9].
Numerous scientific researchers have unveiled various factors that influence the rate of biodegradation. One important requirement is the presence of microorganisms with the appropriate metabolic capabilities, if these microorganisms are present, then optimal rate of growth and hydrocarbon biodegradation can be sustained by ensuring that adequate concentration of nutrient and oxygen is present and that the pH is between 6 and 9 [10]. The physical and chemical characteristics of the oil and the oil surface area are also important determinants of bioremediation success. There are two main approaches to oil spill bioremediation: Bio-augmentation in which known oil degrading bacteria are added to supplement the existing microbial population and Bio-stimulation in which the growth of indigenous oil degraders is stimulated by the addition of nutrient and other growth limiting co-substrate. Nearly all soils and sediments have populations of bacteria and other organisms capable of degrading petroleum hydrocarbons [11]. And in the context of this study, the emphasy is laid on bioremediation through the recovery of appropriate bacterial strains as well as the possibility of the use for microbially enhanced oil recovery for productivity and economic reasons.
Some environmental conditions including temperature and pH can enhance oil degradation by microorganisms. The breaking down of hydrocarbons by bacteria and fungi at the initial steps include the oxidation of the substrates by enzymes oxygenizes in which molecular oxygen is required. The oxygen can be depleted based on its usage [12]. Degradation also occurs in the absence of oxygen. Under anaerobic conditions, some hydrocarbons are degraded or transformed. For example, the anaerobic transformation of benzene and toluene under methanogenic conditions has been characterized as fermentation process in which the substrates are partially reduced, yielding carbon dioxide and methane as end product [13]. The objective of this study is therefore to isolate and identify bacteria which are capable of degrading hydrocarbon from oil polluted soil sample from Ayetoro community in Ilaje Local Government Area of Ondo state. Similarly, the conditions that enhance this degradative process will be beneficial for cleanup purpose in oil polluted areas.
Materials and Methods
Study site and sample collection
The soil samples were collected from an oil-polluted land in Ayetoro community of Ilaje Local Government Area of Ondo state. Sample was collected directly into an aluminum foil paper using sterile spatula, and transported immediately to the Microbiology laboratory of Adekunle Ajasin University, Akungba-Akoko where microbiological analysis was carried out.
Materials and equipment used
The materials including glass wares used for this study were adequately sterilized in the hot air oven at 170 ᵒC for two hours and allowed to cool before use. Similarly, culture medium such as Nutrient agar used was prepared according to manufacturer’s instructions while the mineral salt medium (MSM) was constituted and prepared in the laboratory. Sterilization of the media was done using Autoclave at 15Ibs (121 ᵒC) for 15 minutes.
Preparation of mineral salt medium (MSM)
The mineral salt medium was prepared in the laboratory having the following composition; Na2HPO4.7H2O, 64g; KH2PO4, 15g; NaCl, 2.5g; NH4Cl, 5.0g; yeast extract; and Agar agar in 1000mls of sterile distilled water. Each salt was dissolved in distilled water before mixing. The pH of the solution was adjusted to 6.8. The medium was then sterilized by Autoclaving at 15Ibs pressure (121 ᵒC) for 15minutes.
Enumeration and isolation of bacterial population
Total bacterial population in the oil-polluted soil sample was enumerated and isolated adopting serial dilution and the standard plate counts technique using pour plate method. 10g of the soil sample was measured into a conical flask and 90ml of sterile distilled water was mixed with the sample. The suspension was properly shaken for three hours to homogenize the solution and this served as the stock solution. Ten-fold serial dilution of all the homogenized mixture was carried out using sterile distilled water as diluents. Seven test tubes containing 9ml of distilled water was used for the serial dilution. Aliquots of 1ml from 10-5 and 10-7 dilutions were introduced into duplicated sterile petri dishes and 20ml of molten nutrient agar incorporated with nystatin to suppress fungal growth, were poured into the plates and swirled to allow homogenization. The plates were incubated at 37 ᵒC for 24 hrs after which colonies on the plates were enumerated and sub culturing of bacterial isolates were done to obtain pure culture. Bacterial colonies were picked with sterile inoculating loop and were streaked on freshly prepared nutrient agar plates. The plates were incubated at 37 ᵒC for 24 hrs. Aliquots of 1ml from dilutions of 10-5 and 10-7 were also plated in duplicates on Mineral Salt Agar using the spread plate technique. Nystatin was added to the Mineral Salt Agar to suppress fungal growth. A filter paper saturated with sterile crude oil was aseptically placed on the inside of the inverted Petri dishes and the culture plates were incubated for 14 days at 37 ᵒC. Plates yielding colonies were afterwards enumerated for bacterial isolates.
Identification of the bacterial isolates
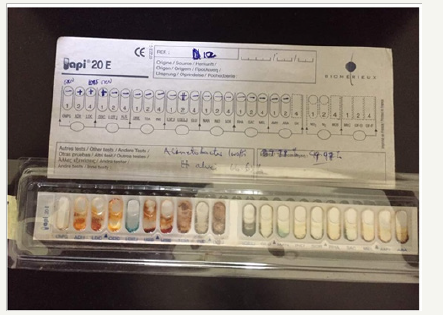
The bacterial isolates were identified by standard microbiological methods. This includes the identification of Gram negative bacteria using API® 20E identification kit.
Growth on bonny light oil
Bonny light oil is a high grade of Nigerian crude oil with low specific gravity, produced in the Niger Delta basin and named after the prolific region around the city of Bonny. The very low sulfur content of Bonny Light crude makes it a highly desired grade for its low corrosiveness to refinery infrastructure and the lower environmental impact of its byproducts in refinery effluent [14].
Method A
Colonies of the different bacterial isolates were transferred into 50ml mineral salts medium (MSM), supplemented with 0.05% (v/v) bonny light oil sterilized by filtration through 0.45 unit membranes (whatmann, USA) and incubated at 28 °C in a water bath shaker at 200rpm for 21 days. Bacterial growth was determined at a 7 day intervals by the physical appearance (i.e. turbidity) and by measuring the optical density (O.D.) at 540nm using Spectronic colorimeter 20 (Bausch and Lomb Inc., Rochester, NY).
Method B
Growth on Bonny light oil was also determined by the ‘holeplate diffusion method’. Twenty milliliters of mineral salts agar medium (MSM) were poured into agar plates. Plates were then inoculated with the test organisms using a sterile swab. Six (6) mm diameter cork borer was used to bore wells on the agar plate and 50 μl of filter sterilized bonny light oil were placed in the holes. Sterile distilled water was used as the control. Agar plates with the bacterial isolates were incubated overnight at 28 ᵒC. The results were recorded after 48 hrs by the physical appearance.
Assay for petroleum oil degradation
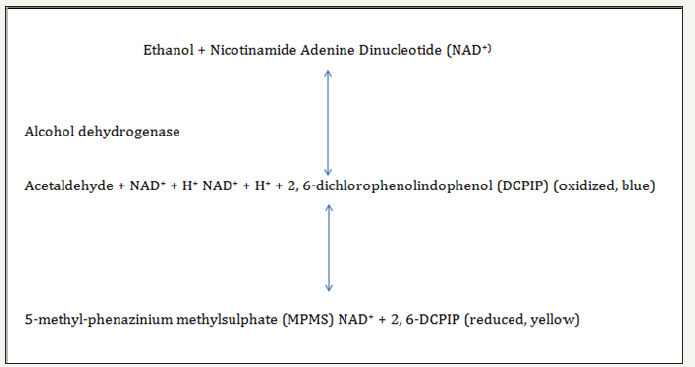
The test was conducted to detect the biodegradation of Bonny light oil, by a monooxygenase biodegradation pathway. The test is based on the following reactions: The test was performed in duplicate at 28 ᵒC in a small test tube containing the following components: 20μl of 0.05mol/l2, 6-dichlorophenolindophenol (DCPIP), 30μ l0.05mol/l 5-methyl-phenazinium methylsulphate (5-MPMS), 25μl of 0.1% (v/v) Bonny light oil, 5μl of 0.15M NAD solution and 25μl of broth containing bacterial cells. Color change was compared visually with four controls. The first control contained no bonny light oil (substrate), the second contained no NAD+ and the third contained no cells. A fourth control consisted of heat-killed cells (10 min at 90 ᵒC). The reaction was followed for 12 hours and examined after 1, 2, 6 and12 hrs.
Results and Discussion
figure 1:Assay for petroleum oil degradation.
This study shows the diversity of microorganisms encountered in soil sample obtained from Ayetoro community, which is part of Ilaje Local Government that is an oil producing Area of Ondo State, Nigeria. The average total bacterial count was 10 x 107cfu/mL. And when cultured on Mineral salt medium (MSM), the average count was 3 x 107cfu/mL. Out of the twenty-two (22) colonies picked at random, (thirteen) 13 were Gram positive while nine (9) were Gram negative (Table 1). The high number of bacteria colonies recovered from the soil sample was due to the organic matter present in the crude oil. This mechanism of oil degradation is as shown in Figure 1 based on the ability of these microbes to utilize petroleum hydrocarbon components for their metabolic activities (Figure 1 & Table 2).
Table 1:The total bacteria count of the soil sample on nutrient agar and mineral salt medium
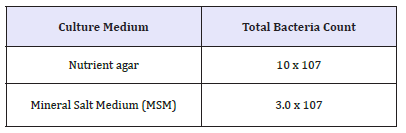
Table 2:Cultural and biochemical characteristics of the Gram positive bacterial isolates.
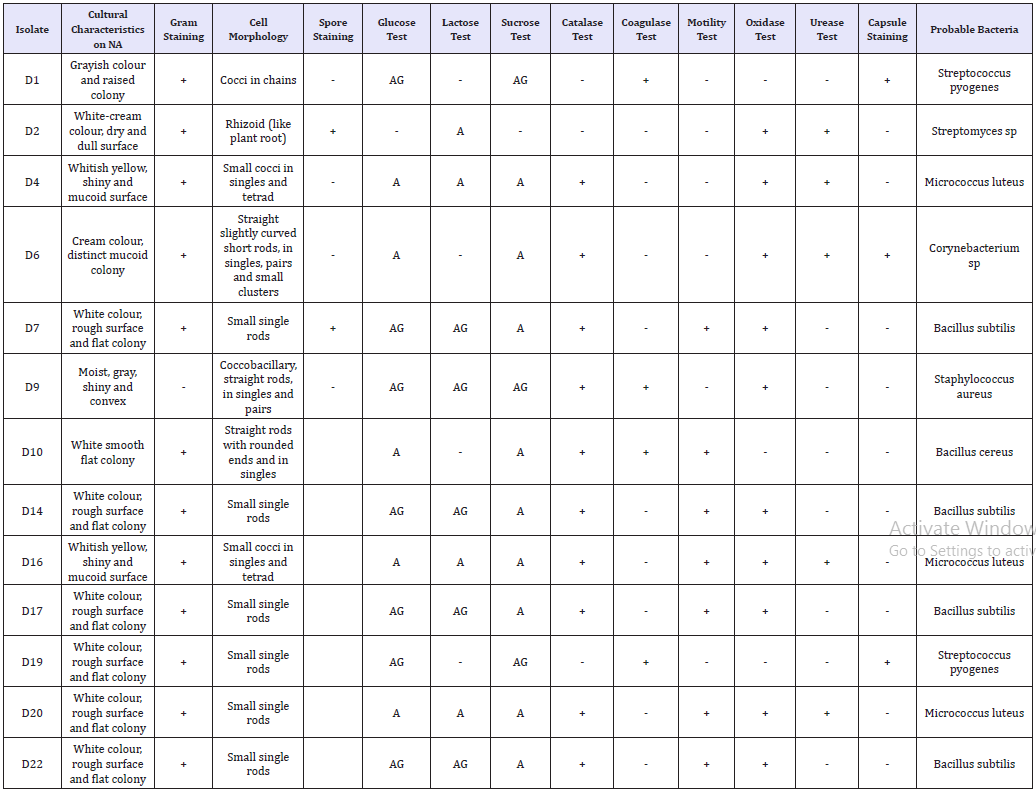
In Table 3, API® identification kit was used for Gram negative bacterial isolates confirmation of identity. The prominence of bacterial load in this ecological zone might have been due to their ability to utilize the organic matter compound present in the crude oil as source of carbon and energy for their metabolic properties. Bacillussubtilis, Bacilluscereus, Pseudomonasaeruginosa, Micrococcus luteus, and Acinetobacter lwoffi obtained in this study were able to degrade Bonny light crude oil while others could not (Table 3). The frequency of oil-degrading bacteria was higher in the soil sample than that of non-oil degrading bacteria. This is likely to be as a result of the metabolic ability possessed by these oil-degrading bacteria such as Bacillus spp and Pseudomonas spp. Which enables the utilization of hydrocarbon present in the crude oil as source of carbon and energy [10]? Similarly, the result obtained in this study is in conformity with the report of [15], who demonstrated that Bacillus spp. and Pseudomonas sp. are of high predominance in hydrocarbon polluted soil as a result of their ability to utilize it.
Table 3:API ® kit identification codes for the gram negative bacterial isolates.
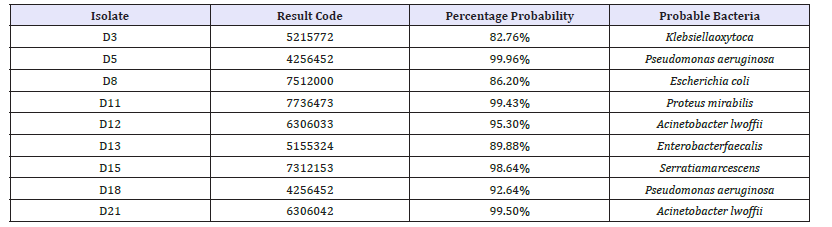
Bacillus spp. produces spores which may shield it from the toxic effects of hydrocarbon. Other organisms such as Streptococcus spp., Klebsiella spp., Proteus spp., and Escherichiacoli were also present in the crude oil polluted soil. Although they may not be able to utilize petroleum products, their presence may be due poor hygienic practices in the community. Microbial degradation of organic contaminants normally occurs as a result of microorganisms using the pollutants for their metabolism, growth and reproduction [16].
The frequency of occurrence of bacteria showed that Bacillusspp had the highest occurrence (22.7%) followed by Micrococcusluteus (13.64%) while Staphylococcusaureus, Klebsiellaoxytoca and scherichiacoli were least in occurrence with value of 4.55% each. This also corroborates the findings of [17], who studied the potential of Pseudomonas sp., Bacillus sp., and Proteus sp. isolated from rivers polluted by hydrocarbons and refinery effluents to degrade petroleum [18]. The result of this investigation showed the occurrence of high numbers of certain oil degrading microorganisms from oil polluted environment as evidence that these microorganisms are the active degraders of such environment [19]. The majority of these organisms isolated were Pseudomonas aeruginosa, Bacillus spp, icrococcusluteus and Acinetobacter lwoffii and the dominance of these organisms has been reported by different researchers as crude oil degraders [5].
plate 1:API kit and typical result of one of the bacterial isolates (Viz: Acinetobacter lwoffi).
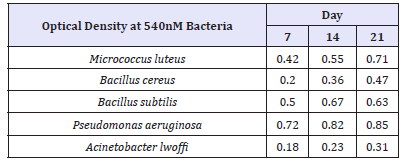
The growth response of different bacterial isolates on Bonny light oil measured as turbidity (O.D. at 540nm) as determined in Table 4. Of the twenty-two (22) bacterial isolates, five (5) showed growth around wells containing Bonny light oil and further showed ability to degrade the oil. The presence of oil-degrading organisms shows that they have adapted to Petroleum-contaminated environment and it is a clear indication that the indigenous microbes were carrying out their metabolic activity. The activities of these microorganisms could be responsible for the bioremediation of the environment and thus clean these polluted sites [5]. Study of microbes that can enhance degradation of environmental pollutants will go a long way to curb the menace of environmental toxicology and protect human health from the surge of diseases spread (Plate 1).
Table 4:The oil-degrading bacteria growth indices (optical density).
Conclusion
The strength of this study entails wide array of bacterial diversity, both Gram positive and Gram negative, that were isolated from oil-polluted environment based on their nature to survive crude oil pollution. These properties will enhance their versatility to be used for cleanup purposes in oil polluted areas. This will make crude oil less harmful to plants and animals activities, thus keeping our environment relatively safe through bioremediation processes. Nevertheless, apart from pollution control through the use of microbe’s encountered, further molecular studies to produce replicate copies of viable organisms for enhanced petroleum recovery purposes is recommended and will be intensified.
References
- Sebiomo A, Bankole SA, Awosanya AO (2010) Determination of the ability of microorganisms isolated from mechanic soil to utilize lubricating oil as carbon source. African Journal of Microbiology Research. 4(21): 2257-2264.
- Kvenvolden KA, Cooper CK (2003) Natural seepage of crude oil into the marine environment. Geo-Marine Letters 23(3-4): 140-146.
- Aboribo RI (2001) Oil politics and the niger delta development commission: the tussle for control and domination. African Journal Environmental Study 2: 168-175.
- Okpokwasili GC (2003) Microbial degradation of petroleum hydrocarbon by brackish water isolates in Nigerian wetlands. In: The Nigerian Man and the Biosphere. Akpata TVI, Okoli A (Eds.), MAB National Committee, USA, pp. 138-146.
- Olukunle OF, Boboye B (2013) The molecular succession of bacterial community of crude oil polluted soil and water samples from the Niger Delta Nigeria. British Journal of Applied Science Technology 3(4): 777- 788.
- Medina Bellver JI, Marní P, Delgado A, Rodrgíuez Sánchez A, Reyes E, et al. (2005) Evidence for In situ crude oil biodegradation after the prestige oil spill. Environ Microbiol 7(6): 773-779.
- April TM, Foght JM, Currah RS (2000) Hydrocarbon-degrading filamentous fungi isolated from flare pit soils in northern and western canada. Can J Microbiol 46(1): 38-49.
- Ulrici W (2000) Contaminant soil areas, different countries and contaminant monitoring of contaminants. In: Rehm HJ, Reed G (Eds.), Environmental process II. Soil Decontamination Biotechnology, (11th edn), pp. 5-42.
- Alfreda ON, Ekene GO (2012) Bioremediation of crude oil polluted soil using bacteria and poultry manure monitored through soybean productivity. Pol J Environ Stud 21(1): 171-176.
- Stephen E, Emmanuel OE, Okpanachi OS, Emmanuel S, Temola OT, et al. (2013) In vitro study of biodegradation of spent lubricating oil by Aspergillusniger. Nature and Science 11(10): 40-44.
- US Environmental Protection Agency (1999) Monitored natural attenuation of petroleum hydrocarbons: US Environmental Protection Agency, Fact Sheet, Office of Research and Development, Washington DC, EPA, USA.
- Bartha R, Bossert I (1984) The treatment and disposal of petroleum wastes. In: Atlas RM (Ed.), Petroleum Microbiology. New York, USA, pp. 553-578.
- Chaillan F, Chayneau CH, Point V, Saliot, Oudot J (2006) Factors inhibiting bioremediation of soil contaminated with weathered oils and drill cuttings. Environ Poll 144(1): 255-265.
- Thomas D (1995) Niger Delta oil production, reserves and field sizes assessed. Oil and Gas Journal 103(6): 18.
- Ijah UJ, Abioye OP (2003) Assessment of physiochemical and microbiological properties of soil, 30 months after kerosene spill. Journal Research Science Management 1(1): 24-30.
- Ghazali FM, Abdul R, Salleh AB, Basri M (2004) Biodegradation of hydrocarbons in soil by microbial consortium. International Biodetermination Biodegradation 54(1): 61-67.
- Okerentugba PO, Ezeronye OU (2003) Petroleum degrading potentials of single and mixed microbial cultures isolated from rivers and refinery effluent in Nigeria. African Journal of Biotechnology 2(9): 288-292.
- Shaopeng Y, Qiuyu W, Lina Q, Cong L (2013) Characterization of oildegrading bacteria from oil-contaminated soil and activity of their enzymes. Biotechnology and Biotechnological Equipment 27(4): 3932- 3938.
- Jain PK, Gupta VK, Gaur RK, Lowry M, Jaroli DP, et al. (2009) Bioremediation of petroleum oil contaminated soil and water. Research Journal of Environmental Toxicology 5(1): 1-26.
© 2018 Ajayi AO. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






