- Submissions

Full Text
Open Access Biostatistics & Bioinformatics
Copulas in Gene’s Relation
Burcu Hudaverdi*
Department of Statistics, Dokuz Eylul University, Turkey
*Corresponding author:Burcu Hudaverdi, Department of Statistics, Dokuz Eylul University, Izmir, Turkey
Submission: May 13, 2023;Published: May 25, 2023

ISSN: 2578-0247 Volume3 Issue3
Opinion
Genes are considered as segments of DNA that provide instructions for the production of specific proteins or RNA molecules. Genes can be related to one another in a variety of ways. These relationships can play important roles in the regulation of cellular processes and functions, as well as in the development and evolution of organisms. Gene relation refers to the relationship between two or more genes in terms of their function, location, or sequence similarity. In studying the expression and relationship of genes, researchers can get a sense of how gene-information is used to know cell functions and how changes in gene expression or regulation can lead to disease. As an example, co-expression analysis can help to determine gene-groups that are functionally related and may be involved in specific biological processes or diseases. Also, network analysis can be used to identify regulatory relationships between genes as well as to build gene regulation networks that control specific cellular processes. See also Allocco et al. [1], Elise et al. [2], Ruan et al. [3] & Song et al. [4], By interacting with each other, genes form a complicated network. The relationship between groups of genes with different functions can be represented as gene networks. Since the accessibility of the large gene expression data, several methods have been developed to analyze the gene-to-gene relationship. Copula approach is being used to study the relationship between the expression profiles of pairs of genes, since a copula is a multivariate model that can be used for explaining gene-gene dependence.
Copulas are mathematical functions that describe the dependence structures between random variables, while allowing for the specification of their marginal distributions separately. They can provide a way to model the dependency structure between variables without making assumptions about the underlying distribution of each variable. In other words, copulas separate the marginal distributions from the dependence structure, allowing for more flexibility in modeling. Sklar [5] showed that a copula exists for any multivariate distribution, such that the joint distribution equals the copula applied to the marginal distributions. Let X1, . . ., Xd be the random variables with joint distribution function H(x1, . . ., xd) with marginals F1(x1)=P(X1 x1),≤ . . .,Fd(xd)=P(Xd xd)≤. The relation between the copula and the joint distribution function of these random variables can be defined as
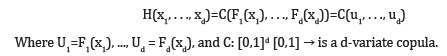
Copulas can be used to model various types of dependence structures, such as positive or negative dependence, asymmetric dependence, or tail dependence. They can be easily extendible to use in the measurement of dependence structure of multiple genes. Recently, some studies demonstrated the usefulness and importance of copulas in gene expression analysis, providing a powerful tool for exploring dependence relationships between genes and identifying key drivers of disease progression. Kim [6] analyzed the gene interactions for two gene data sets through directional dependence by copula approach. Emura & Chen [7] proposed using copulas to model the dependence between censoring times. To address the issue of dependent censoring, they demonstrate the copula-based procedure for selecting genes, developing a predictor, and validating the predictor using non-small-cell lung cancer data. Ray [8] studied a copula-based framework to model differential co-expression between gene pairs in two different conditions. Sun & Ding [9] proposed a novel class of copula-based semiparametric transformation models for bivariate data under general interval censoring.
The advantage of using copulas in gene relation analysis is that they provide a flexible and powerful way for modeling the complex dependence structure between genes, which can be difficult to capture using traditional correlation or regressionbased approaches. They can model nonlinear and non-monotonic dependency between genes. This is important in gene relation analysis, because the relationship between genes may not always be linear or monotonic. For example, the expression of one gene may only be related to the expression of another gene within a certain range of expression levels. Copulas can also be used to model the joint distribution of multiple genes, allowing researchers to identify groups of genes that are co-expressed or functionally related. By identifying these groups of genes, researchers can gain insight into the biological processes that are regulated by these genes, which can be useful for understanding the underlying mechanisms of disease. Overall, copulas in gene relation analysis can be used to identify co-expressed genes, construct gene networks, and investigate the relationship between gene expression and other variables of interest.
References
- Allocco DJ, Kohane IS, Butte AJ (2004) Quantifying the relationship between co-expression, co-regulation and gene function. BMC Bioinformatics 5: 18.
- Serin EAR, Nijveen H, Hilhorst HWM, Ligterink W (2016) Learning from co-expression networks: possibilities and challenges. Front Plant Sci 7: 1-18.
- Ruan J, Dean AK, Zhang W (2010) A general co-expression network-based approach to gene expression analysis: Comparison and applications. BMC Syst Biol 8: 4.
- Song L, Langfelder P, Horvath S (2012) Comparison of co-expression measures: mutual information, correlation, and model-based indices. BMC Bioinformatics 13: 328.
- Sklar A (1959) Markov-switching time-varying copula modeling of dependence structure between oil and GCC stock markets. Distribution Functions in N Dimensions and their Margins, Publications of the Institute of Statistics of the University of Paris, France 8: 229-231.
- Kim JM, Jung YS, Sungur EA, Kap HH, Changyi P, et al. (2008) A copula method for modeling directional dependence of genes. BMC Bioinformatics 9: 225.
- Emura T, Chen YH (2018) Gene selection and survival prediction under dependent censoring. In: Analysis of survival data with dependent censoring. Springer Briefs in Statistics, pp. 75-84.
- Ray S, Lall S, Bandyopadhyay S (2020) CODC: A copula-based model to identify differential co-expression. NPJ Systems Biology and Applications 6: 20.
- Tao S, Ying D (2021) Copula-based semiparametric regression method for bivariate data under general interval censoring. Biostatistics 22(2): 315-330.
© 2020 Burcu Hudaverdi. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






