- Submissions

Full Text
Novel Techniques in Nutrition and Food Science
Molecular Methods of Testing Tuberculosis Resistant to Antibiotics in Patients Infected in Indore, M.P India
Deva Rupal*
Oncquest Laboratories ltd., India
*Corresponding author: Deva Rupal, Oncquest Laboratories ltd., India
Submission: May 14, 2018;Published: November 13, 2018

ISSN 2640-9208 Volume3 Issue1
Abstract
Objective: Our objective of the work was to identify the type of Mycobacterium at species level with an aim to diagnose and treat the resistant strains by using MDR strains in infecting in patients to identify the infecting Mycobacterium and to find resistance at the level of genes tested using molecular biology of line probe assay methods.
Design: We tested various samples for Mycobacterium by using microbiology and molecular biology method of TB-TMA from clinical suspects. With use of Amplification of gene and line arrays of bacteria we determined the susceptibility character of drugs of infecting Mycobacteria in the patients.
Results: We were able to identify Mycobacterium tuberculosis by measuring the activity at Promoter level of Mycobacteria and determined their first and send line of drug sensitivity using molecular biological tools of drug susceptibility of MDR and XDR sure methods developed by us. We could detect mutations in repo B gene imparting drug resistance to Rifampicin or Kat G gene and In A gene to Isoniazid from patients’ samples infected with Mycobacterial species or resistance to fluoroquinolone, by detecting mutations in gyrA gene and rrs gene that imparts resistance of bacteria to injectable antibiotics. We were able to also Identify Mycobacterium Other Then Tuberculosis (MOTT)
Material and methods
Samples: We obtained samples of clinical TB suspects from various multispecialized hospitals in our Norquest laboratory at Central lab, Indore (M.P). These patients were recorded in the hospitals and tested using medical, for treatment of patients. The clinical suspects were tested by medical specialists to have pulmonary or extra pulmonary TB, tested by medical methods like chest radiography, C.T scan, pleural tap and CBC tests and dispatched. The samples were transported immediately to our laboratory under cold temperature conditions. Both the pulmonary as well as extra pulmonary samples were analyzed in our laboratory for diagnosis of tuberculosis. For Drug. Sensitivity test three consecutive early morning sputum samples were collected by laboratory technologist, according to protocol in our NABL accredited, Central lab-Norquest, Indore. The samples were processed on the same day or were kept at +4ᵒC till processed. The Medical diagnosis of suspects was done by medical specialists. And M. tuberculosis was detected using TB-TMA method to identify the infecting bacteria as described earlier.
Conclusion: We found great variations in the genes of Mycobacterium infecting patients tested in our lab. We were able to find drug sensitivity or resistance of each bacteria infecting, which proved out to be very useful for Clinicians for prescription of the drug regimen based on the infecting bacteria in individual patient according to type of species the patient is infected with Multidrug-resistant tuberculosis (MDR-TB) is tuberculosis disease caused by organisms, which show different level of resistance to Isoniazid or Rifampicin, which may not show resistance to other anti-TB drugs. With increase in MDR TB around the world, among new cases, as well as in the previously treated ones as reported by WHO (2010) [1] & [2].
Introduction
The condition that occurs due to usage of antibiotics, by patient, who halts the medicine ragmen upon feeling better or due to lack of affordable drugs sufficient to kill 100% bacteria becoming resistant, to even to the two most powerful anti- Mycobacterium drugs like Isoniazid (INH) and Rifampicin (RMP). The first-line treatment anti-TB drugs may result in further spread of MDR bacteria in the Population [3]. Utilizing molecular detection of tuberculosis from clinical suspects in our study included samples presented here from various tissues for isolation of Mycobacterium to study the drug susceptibility of each bacteria infecting patient isolated from clinical suspects in our laboratory.
Earlier, we had reported utilization of modern molecular biology methods in conjugation with classical methods for identification of M. tuberculosis infection to assist with the judgment for prescription of right therapy for the Mycobacterium infected individual [3], The prevalence of pulmonary tuberculosis in district Jabalpur were reported [4]. We utilized the, methods of identified by Mycobacterium, using modern molecular biology tools and recording the occurrence of multi drug resistance in Mycobacterium from the isolates from clinical suspects obtained from our various centers of India, in our lab. We in this short report identified the occurrence of MDR TB existence in India and presented. India is among 27 MDR-TB countries, the country holds a huge burden of MDR TB and, holding high MDR-TB [3,5]. We present herewith the type of infection bacteria in infected patients using molecular identification of bacteria at their genetic level for picking the drug of choice for treatment in patient.
Result
Figure 1:First line drug resistance-Line array for identification of drug sensitivity/resistance in Mycobacteria by identification of rpo, inh A, kat G gene in Mycobacteriu. tuberculosis.
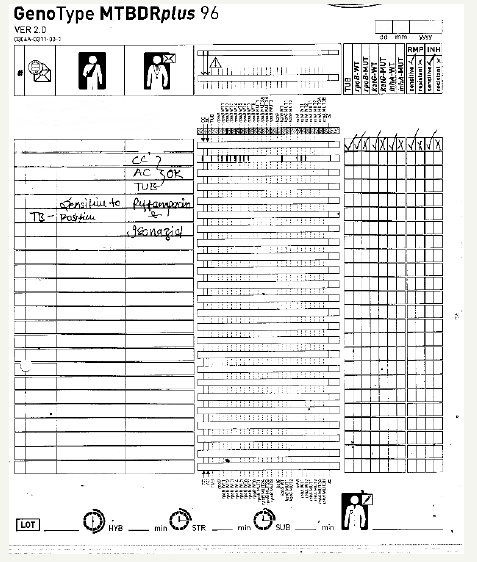
Figure 2:2nd line drug resistance- Line array for identification of Drug sensitivity/resistance in Mycobacteria with identification of ggrA, rrs and embB gene in Mycobacterium tuberculosis.

Using molecular biology-based line array we were able to detect Mycobacterium anti biotic sensitivity. As seen in Figure 1, the infecting bacteria found was found to be resistant to first line drug Isoniazid and Rifampicin. The control lines of Conjugate control (CC), Amplification (AC) and Mycobacterium tuberculosis (TUB) were detected there was clear mark on wild type bands (WT1-7) were detected, rpo B marking as noted in Figure 2. demonstrating the and Kat G WT indicating sensitivity to both Rifampicin and Isoniazid drugs by line line array for Mycobacteria, while no resistant gene bands were found.
Figure 3:A identification of MOTT by Analysis Gc- rich gram-positive bacteria.

Figure 4:Identification of MOTT - M. kanasaii.
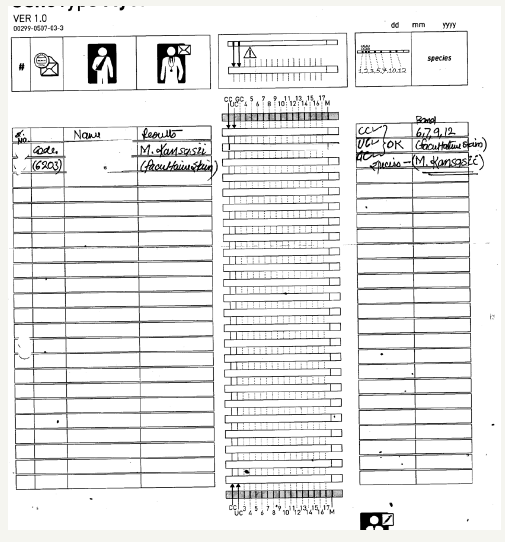
In one patient test for second line of drugs sensitivity infection with isolate resistant to fluoroquilone by using molecular biology, detected isolate resistant to fluproquazone, though showed hybridized line for gyr A wild type (WT) were seen as well as Gyr A mutant and with line for rss gene Wild type were detected and EmbB gene ethambutol and injectable antibiotics like kanamycin, amikacin and capreomycin and viomylin as seen in Figure 3. Another infected patient, we were able to detect hi GC rich bacteria as seen in Figure 4, and detected patient infected with facultative strain of M. kansasii as seen in Figure 4 and could detect M. chelonae as seen in Figure 5. The control bands CC, AC and TUB bands were found to be up to the mark Indicating correct mark for the test performed.
Figure 5:Identification of MOTT -M. chelonae.

Discussion
The World Health Organization has now in year 2016 has announced a new recommendation for a nine-month shortened treatment regimen for multi-drug resistant tuberculosis (MDR-TB) patients. Years of research and clinical studies from The Union and its partners have proved fundamental in preparing an evidence base for this ground-breaking announcement and World Health Organization has accepted the rapid method of line probe assay recently towards the treatment of MDR patients. Earlier followed treatment against MDR-TB had included, which had terrible side effects, including permanent hearing loss. The treatment is tedious for clinicians to administer along with patients to follow regimen strictly. Globally MDR-TB is a public health emergency where, 480,000 people contract MDR-TB are found which are further rising. This is the reason on-going studies into even shorter regimens to continue further. Diagnostic Research and evidence are fundamental to beating the tuberculosis disease.
Presence of a resistant strain of M. tuberculosis in a patient newly diagnosed with TB who were not previously been treated with anti TB drugs along with the old patients who were earlier treated with TB Therapy medication. Such treated patients are likely to have been infected with a strain that was already drug resistant. Such cases may possess “primary” drug resistance. The drug resistance in a previously treated TB case is the presence of a resistant strain in a TB patient who has previously received at least one month of TB therapy, but during the course of antituberculosis treatment, drug resistance could have emerged as “secondary” or “acquired” drug resistance. Thus, tuberculosis epidemiology is globally becoming a complex blend of reactivation of organism, leading to further spread of newly developed strains, in the population. Close molecular biology-based investigations are helpful for deciding the strategies to identify and treat the developed of infection in the population. For induction of therapy to be provided to infected patients to stop further spread accordingly. This type of study is useful for understanding the development of infection in a population to assist and to direct the patient towards the therapy. Understanding the occurrence and spread of infection in Community is crucial for the management of Drug-Resistant TB. which is though difficult and should not be over looked? The treatment of drug resistant TB depends upon the bacterial agent infecting the patient for example the patient is treated with regimen for 2 months followed by 4 months of rifampin and Isoniazid as described earlier [4].
Previously, we had reported utilization of modern method of molecular biology in conjunction with classical methods for diagnosis of M. tuberculosis, for assisting clinicians for their quick decision for directing patients for perception to drugs [4]. While Dr. Bajpai [6] had highlighted the importance of combination of AFB smear microscopy, culture, histopathology and PCR methods for diagnosis of genital tuberculosis in women clinical suspects. In present work, apart from identification of the Mycobacterium sp, we could detect Mycobacterium, at their species level or detecting Mycobacterium other then tuberculosis, to know their antibiotic susceptibility. We tested both pulmonary and extra pulmonary samples, from clinical suspects of TB for detection of Bacteria at gene level and determined the Multiple Drug Resistance in Mycobacterium, occurring in, in our laboratory. We were able to identify type of MDR Mycobacterium detected for choosing the ideal therapy for individual patent, depending upon the infecting microorganism. Mostly all Mycobacteria drug resistant species can be controlled by drugs and/or resection surgery [7]. The regimens for treating M. tuberculosis have an initial phase of 2 months, followed by a either of several options for the continuation phase treatment in patients of either 4 or 7 months, under strict rule of non-interruption of drug usage. The treatment of drug resistant bacteria is carried out with the usage of combination of drugs that augment effect of individual drug for the treatment of Multi Drug-Resistant tuberculosis in a positive way [8]. We had recently studied multi drug resistant pattern in samples of clinical suspects from Indore [9]. Treatment of patients depends depending upon the identified tuberculosis as described before [10]. In today’s era of development of new multi drug resistant Mycobacterium, in country new therapeutic regimen is needed to be able to completely destroy the infecting resistant Mycobacterium, as reported earlier [11- 14]. The Mycobacterium tuberculosis complex (MTBC), have been reported to vary widely in different countries [14]. In the present study presented herewith we present the findings of detection of Mycobacterium infecting in India, tested in our laboratories [15-20].
That in India. MDR-TB is still at a higher risk and can be further amplified to develop newer strains along with preexisting strains which could be associated with development of extreme drug resistant (XDR-TB) Mycobacterium may there by further prorogate in the population if not controlled [21]. Our data for Molecular biology methods of identification of type of bacteria infecting patients indicates towards controlling infection spread further in the population in India. In our tests we could visualize conjugate control band (CC) that determines the efficiency of conjugate binding and substrate reacting as and TUB band determining M. tuberculosis Complex shown in Figure 1. To find Inh and rpo B wild type genotype as seen in Figure 1 directing towards the infection with no resistance band detected pointing towards the bacteria to be sensitive to both rifampicin and isoniazid. M. tuberculosis complex include pathogens causing the classical M. tuberculosis, M. bovis, M. afri-canum,M. microti, as well as the newly recognized MTBC members, M. pinniped and M. caprae species [22- 24]. Even with evolution, M. tuberculosis remains one of the most established pathogens known to humans inspire the availability of its treatment, vaccine and antimicrobial agents. The adaptability of Mycobacterium reflects a very ancient mark of evolution. The method we used include specific DNA extraction from bacteria from clinical sample, followed by amplification with biotinylated primers and further reverse hybridization of single stranded biotin labeled amplicons to membrane bound probes and detecting the developed with addition of a streptavidin/ alkaline phosphates conjugate as described in Material and methods [25]. We used MTBDR plus test based on DNA-Strip to identify M tuberculosis complex and its resistance to Rifampicin or isoniazid from patient material. The identification of rifampicin was identified by mutation of rpoB gene [26-28]. While high level Isoniazid was detected by ket G or inh A located in promoter region of Mycobacterium. In most patients we detected sensitivity as shown in Figure 2. We were able to detect Mycobacterium resistant to fluoroquilones (e.g.ofloxacin and moxifloxin) showing mutation in gyrA gene, and rrs gene, but resistant to injectable antibiotics but sensitive to ethambutol’s with no occurrence of mutations in embB gene as shown in. We were able to detect. infection due to Mycobacteria other than tuberculosis complex, by different species including M. kansasii, M. chelonae, M. immumogenum infection as seen in Figure 3-5 at gene level of infecting Mycobacteria in year 2012- 2014. The, we present herewith the work of diagnosis done in our Norquest Indore laboratory with the use of modern molecular biology methods [29- 33].
Conclusion
We were able to detect different variations in the genes of drug resistant Mycobacterium infecting patients obtained from our various centers in India. We were able to find drug sensitivity of each bacteria infecting which proved out to be very useful for Clinicians to specifically to prescribe the drug regimen based upon the infecting bacteria in each patient.
References
- WHO (2010) Executive summary multidrug and extensively drugresistant TB (M/XDR-TB). 2010: 1-3.
- Ajay H, Ruby J, Vijayselvi R (2014) Maternal mortality and derivations from the WHO near-miss tool: An institutional experience over a decade in Southern India. J Turkish Ger Gynecol Assoc 15(4): 222-227.
- Ormerod LP (2005) Multidrug-resistant tuberculosis (MDR-TB): Epidemiology, prevention and treatment. Br Med Bull 73-74: 17-24.
- Deva R, Songara P, Neema S, Kothari V, Mehra A, et al. (2014) An interdisciplinary parallel methodological approach for the diagnosis of tuberculosis in the Central state of India. 13(9): 70-77.
- World Health Organization (2013) Global tuberculosis report.
- Srivastava I, Bhatambare GS, Deshmukh AB, Bajpai T (2014) Original research article genital tuberculosis: Evaluating microscopy, culture, histopathology and PCR for diagnosis all play their role. 3(4): 439-445.
- Goel G, Khatuja R, Radhakrishnan G, Agarwal R, Agarwal S, et al. (2013) Role of newer methods of diagnosing genital tuberculosis in infertile women. Indian J Pathol Microbiol 56: 155-157.
- Ameeruddin NU, Luke EH (2014) Impact of isoniazid resistance on virulence of global and south Indian clinical isolates of Mycobacterium tuberculosis. Tuberculosis (Edinb) 94(6): 557-563.
- Prafulla S, Rupal D, Ila S Bajpai, Sushma N, Vinita K, et al. (2015) Drug resistance patterns of Mycobacterium tuberculosis - isolates from indore, India. British Journal of Medicine & Medical Research ISSN: 2231-0614: 10(5).
- Nachega JB, Chaisson RE (2003) Tuberculosis drug resistance: a global threat. Clin Infect Dis 15(36): S24-30.
- Neil M, Schluger W. Diagnosis, treatment, and prevention of drugresistant tuberculosis.
- Chang KC, Leung CC, Yew WW, Leung E, Leung WM, et al. (2012) Pyrazinamide may improve fluoroquinolone-based treatment of multidrug-resistant tuberculosis. Antimicrob Agents Chemother 56(11): 5465-5475.
- Jain K, Desai M, Solanki R, Dikshi RK (2014) Treatmen0074 outcome of standardized regimen in patients with multidrug resistant tuberculosis. J Pharmacol Pharmacother 5(2): 145-149.
- Bonura C, Georgiou MK, Refrégier G, Aleo A, Fasciana T, et al. (2014) Molecular epidemiology of tuberculosis in Sicily, Italy: what has changed after a decade. BMC Infect Dis 14(1): 602.
- Winn WC, Allen SD, Janda WM, Koneman EW, Schrecken PC, et al. (2009) Koneman’s color atlas and textbook of diagnostic microbiology. LWW, (6th edn), Lippincott Williams & Wilkins, New York, USA.
- Rozati R, Roopa S, Rajeshwari CN (2006) Evaluation of women with infertility and genital tuberculosis. 56(5): 423-426.
- Goel MM, Budhwar P, Jain A (2012) Immunocytochemistry versus nucleic acid amplification in fine needle aspirates and tissues of extrapulmonary tuberculosis. J Cytol 29(3): 157-164.
- Government of India (2013) TB India 2013: revised national tb control programmed-annual status report.
- Genç SNB, Solak A, Mayda A (2013) Isolated tuberculous tenosynovitis of the anterior tibial and extensor digitorum longus tendons. J Clin Imaging Sci 3(37).
- Kimizuka HNY, Ishii M, Murakami K, Ishioka K, Yagi K et al. (2013) A case of skeletal tuberculosis and psoas abscess: disease activity evaluated using 18 F-fluorodeoxyglucose positron emission tomographycomputed tomography. BMC Med Imaging 13(1): 37.
- Kalra N, Agrawal P, Mittal V, Kochhar R, Gupta V, et al.
- Lee VY, Wong JT, Fan HC (2012) Tuberculous pericarditis presenting as massive hemorrhagic pericardial effusion. BMJ Case Rep.
- Ahmad ANF, Huda N (2013) Osteoarticular tuberculosis-a three years’ retrospective study. J Clin Diagn Res 7(10): 2189-2192.
- Narayana Reddy RA, Narayana SM, Shariff S (2013) Role of fine-needle aspiration cytology and fluid cytology in extra-pulmonary tuberculosis. Diagn Cytopathol 41(5): 392-398.
- Boubaker K, Gargah T, Abdulrahim E, Abdallah TB, Khader A (2013) Mycobacterium tuberculosis infection following Kidney Transplantation. Biomed Res Int p. 347103.
- Cho YS, Joo KJ, Kwon CH, Park HJ, (2013) Tuberculosis of testis and prostate that mimicked testicular cancer in young male soccer player. J Exerc Rehabil 9(3): 389-393.
- Scheepers MA, Lecuona KA, Rogers G, Bunce C, Corcoran C, et al. (2013) The value of routine polymerase chain reaction analysis of intraocular fluid specimens in the diagnosis of infectious posterior uveitis. Scientific World Journal p. 545149.
- Mehta S (2013) Clinical utility of 18 Fluorodeoxyglucose (FDG)-PET/CT scans in patients with suspect ocular tuberculosis. Indian J Ophthalmol 61(10): 603-605.
- Rao VG, Bhat J, Yadav R, Gopalan GP, Nagamiah S, et al. (2012) Prevalence of pulmonary tuberculosis-a baseline survey in central India. PLoS One 7(8): e43225.
- M. & McCartney (1996) Pract Med Microbiol Churchill Livingstone, (Elsevier).
- Manca C, Paul S, Iii CEB, Freedman VH, Kaplan G (1999) Mycobacterium tuberculosis catalase and peroxidase activities and resistance to oxidative killing in human monocytes in vitro. Infect Immun 67(1): 74- 79.
- Dixit P JA, Singh U, Sharma P. J Microbiol Methods 88 (1): 122-126.
- Hemvani NCDS, Patidar V, Chitnis DS (2012) In-house, simple & economical phage technique for rapid detection of rifampicin, isoniazid, ethambutol, streptomycin & ciprofloxacin drug resistance using Mycobacterium tuberculosis isolates. Indian J Med Res 135(5): 783-787.
© 2018 Deva Rupal. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






