- Submissions

Full Text
Novel Research in Sciences
On the Mechanism of Antibiotic Production in Co-Cultures of Generated DNA (Streptomyces Griseus) Crown Cells (Artificial Cells) with Yeast (Beer)
Shoshi Inooka1*
1Japan Association of Science Specialists, Japan
*Corresponding author: Shoshi Inooka, Japan Association of Science Specialists, Japan
Submission: December 08, 2020;Published: December 16, 2020
.jpg)
Volume5 Issue3December, 2020
Abstract
DNA crown cells are artificial cells in which the outside of the membrane is coated with DNA. DNA crown cells (artificial cells) can be generated by incubating cells within egg white containing a sphingosine (Sph)-DNA-adenosine mixture. Previous research investigating the contribution of DNA crown cells to applied fields demonstrated that antibiotics are produced by co-cultures of yeast (beer) with DNA crown cells when the latter are generated using DNA from the antibiotic-producing Streptomyces griseus. However, many problems remain in the characterization of these phenomena. Notably, there is a long delay between the initiation of the co-culture and antibiotic production, either about 5 weeks when using DNA crown cells in egg white or about 15 days when using purified cells.
The present experiments were carried out to resolve the mechanism of antibiotic production, especially the issue of the delay in antibiotic production in co-culture systems. The present study demonstrated that antibiotic was produced after 5 days of co-culturing at small scale (1mL malt) and that the yeast was unable to grow once antibiotic production initiated. Based on these results, I propose the following mechanism for antibiotic production in this co-culture system. Time until antibiotic was detected may be influenced by the volume of malt used and the ratio of contact between the DNA crown cells and yeast. Notably, antibiotic was detected after 5 days of culturing at small scale (using 1mL malt and cells incubated with yeast (3mg) in 2mL egg), after 17 days at intermediate scale (using 30mL malt and cells incubated with yeast (0.3g) in 6mL egg white), and after 5 weeks at large scale (using 10L malt and cells incubated with yeast (5g) in 20mL egg white).
Yeast that were in contact with DNA crown cells produced antibiotic, but were unable to grow after the initiation of antibiotic production. The remaining yeast that were not in direct contact with the DNA crown cells continued to grow and produce antibiotic and to accumulate the antibiotic. Further experiments will be needed to validate this proposed mechanism. With or without such validation, there remain several problems that will need to be addressed, including whether DNA crown cells in fact are killing the cocultured yeast
Keywords: Antibiotic;Beer; DNA crown cells;Streptomyces griseus;Yeast
Introduction
Several approaches have been reported for generating fully operational (self-replicating) artificial cells [1,2]. In one approach, such artificial cells are covered with DNA (yielding a cell type called DNA crown cells) [3] and incubated in egg white. In theory, unlimited classes of DNA crown cells could be prepared; in practice, the method has been used to generate several kinds of DNA crown cells [4-7]. To clarify whether such DNA crown cells can contribute to fields such as biomedicine or bio-industry, experiments were performed demonstrating that antibiotic is produced by co-cultures of yeast (beer) with DNA crown cells covered with DNA from the antibiotic-synthesizing Streptomyces griseus [8]. Previous experiments have suggested that antibiotics are produced from the beer in such co-culture systems. However, such co-culture systems present several problems that must be resolved to facilitate contribution in the relevant fields of applied science.
One of these challenges is length of time required before antibiotic production is obtained. For example, antibiotic production takes 5 weeks for co-culture in 10L of malt with egg white [8] or 17 days for co-culture in 30mL of malt with purified cells [9]. The experiments described here sought to clarify the mechanism of antibiotic production, especially with respect to the time required to achieve antibiotic production. Notably, I demonstrated that antibiotic production was obtained in 5 days with small-scale co-cultures of yeast and DNA crown cells in 1mL of malt, but the initiation of antibiotic production resulted in the inhibition of yeast growth (and possibly yeast cell death). Based on these findings, we propose a mechanism for antibiotic production by co-cultures of DNA crown cells and yeast.
Materials and Methods
Materials
The following materials were used:
A. The preparation of DNA crown cells employed the following materials:
Sphingosine (Sph) was obtained from Tokyo Kasei (Japan). Adenosine was obtained from Wako (Japan). DNA was extracted from Streptomyces griseus. Monolaurin was obtained from Tokyo Kasei (Japan). Edible White Leghorn eggs were purchased from a local market in Japan. The A-M compound was synthesized as mixtures of adenosine and monolaurin as described previously [10].
B. Co-cultures employed the following materials:
Yeast was obtained as Dry Ale Yeast (Safale S-04) from Fermentis Bergy. The malt medium was obtained as Black Rock Pilsner (New Zealand). These materials were used in combination with an AUBERCRAFT (Okazaki, Japan) handmade-beer kit, as described in the kit instructions. Dulbecco’s Minimal Essential Medium (D-MEM) was purchased from Sigma-Aldrich. Complete medium consisted of D-MEM supplemented with 10% bovine serum (FBS; Sigma-Aldrich,USA)
C. Antibiotic production testing employed the following materials:
The medium consisted of Potato Dextrose Agar (PDA), purchased from Kyodo-Nyugyo (Tokyo, Japan). The testing bacterial strain was Bacillus subtilis natto, obtained as a freeze-dried stock from Daikokuya (Nagoya, Japan) and suspended at 10 mg/mL in sterile distilled water prior to use in testing.
Methods
Preparation of DNA (S. griseus) crown cells : Artificial cells were generated using Sph-DNA-A-M as described previously [8]. Briefly, Sph and DNA were mixed, and the mixture then was heated. A-M compound was added. After injection of the mixture, eggs were incubated at 37 °C for 7 days. The egg white then was recovered from the injected eggs and combined with yeast.
Separation of DNA crown cells from egg white : In the present experiments, DNA crown cells were purified using D-MEM as described previously [3].
Preparation of sample (beer) to test growth of yeast: The first experiment was performed as described previously [9]. Briefly, dry yeast (1g) and DNA crown cells that were purified from 6mL of egg white in 4 generations (four cycles of passage from one egg from the next) were mixed and incubated for 5 hours at 37 °C. Next, the yeast-DNA crown cell mixture (1mL) was combined with 30mL of beer malt, and the mixture was incubated for 18 days at room temperature. The resulting co-culture then was examined for the production of antibiotic and for the cultivation of yeast.
Preparation of sample (beer) to assay antibiotic in small-scale co-culture: In the second experiment, dry yeast (3mg) and purified DNA (S. griseus) crown cells purified from 2mL egg white were combined with 1mL of D-MEM complete medium, and the mixture was incubated for 5 hours at 37 °C. Next, the resulting mixture was distributed at 50µL/tube to three microfuge tubes containing 1mL malt each; the tubes then were incubated for 5 days at room temperature. The resulting spent medium was used to test antibiotic activity. Next, the malt from the three tubes incubated 5 days at room temperature was sub-cultured. Specifically, the malt was distributed at 50µL/tube to five microfuge tubes containing 1mL fresh malt each; the tubes then were incubated for 5 days at room temperature. The resulting spent medium was used to test antibiotic activity.
Microscopic observation and cultivation of samples: The yeast in malts of the first experiment were spotted to glass slides and observed by light microscopy. Additionally, 1mL of each of the same samples were cultured for 7 days at room temperature using Compact DryR YM medium (Nissui, Japan) according to the manufacturer’s instructions.
Preparation of plate to assay antibiotic production: The antibiotic assay was carried out using the agar well method, as described previously [8]. Aliquots (5mL) of the test bacteria were mixed with 200mL agar. Aliquots (approximately 15mL/plate) of the resulting bacteria-agar mix were dispensed into Petri plates. Following solidification, a well of about 2cm in diameter was generated in each plate. The test fluid (400µL/sample) was dispensed into each well (each dish), and the plates were incubated for 18 hours at 37 °C. After incubation, the size of the inhibition zone was observed.
Results and Discussion
Antibiotic production and cultivation of yeast in co-cultures
To investigate whether DNA crown cells might contribute to applied science, I performed co-culture of yeast with DNA crown cells prepared in the presence of DNA from S. griseus (designated DNA (S. griseus) crown cells). S. griseus produces both chromomycin and streptomycin; the strain of yeast was one employed for beer fermentation. The co-culture system was assessed for the production of antibiotics. Notably, similar systems have presented several problems for antibiotic production in previous experiments. One of these challenges was the length of time required to achieve antibiotic production. For example, previous experiments showed that antibiotic synthesis takes 5 weeks in co-cultures of yeast and egg white-grown DNA (S .griseus) crown cells, and 15 days in co-cultures of yeast with purified DNA (S. griseus) crown cells. Therefore, the present study sought to resolve why it takes such a long time to achieve antibiotic production in such systems.
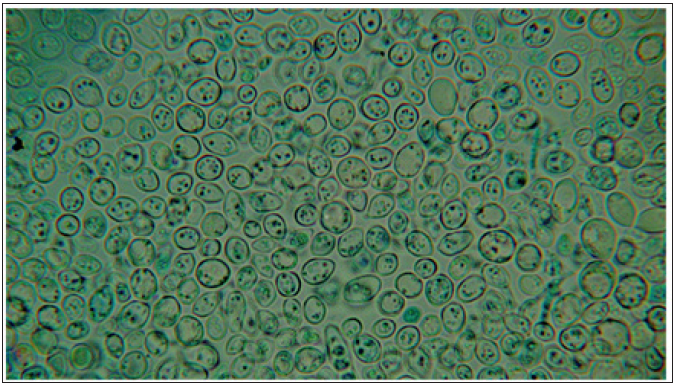
In the first experiment, the co-cultures of yeast with DNA crown cells were carried out according to the previously described methods. The results (data not shown) confirmed that antibiotics were produced after 18 days of co-culturing. Next, 1-mL aliquots of the resulting malts were combined with yeast growth medium (Compact DryRYM), and the mixtures were incubated for 3~7 days at room temperature. No colonies were observed (Figure 1a & 1b), suggesting that these yeast were no longer viable. In parallel, a drop of the same sample (malt) was placed on a glass slide and observed by light microscopy. The presence of yeast with standard morphology was confirmed (Figure 2), but these cells were not further characterized.
Figure 1: The appearance of plates after cultivating the malt from 18-day co-cultures of yeast with DNA crown cells, conditions that were shown to result in antibiotic production. No colonies were observed, suggesting that any remaining yeast were unable to grow. Figure 1b: Culture of yeast as a control. Blue colonies were observed.
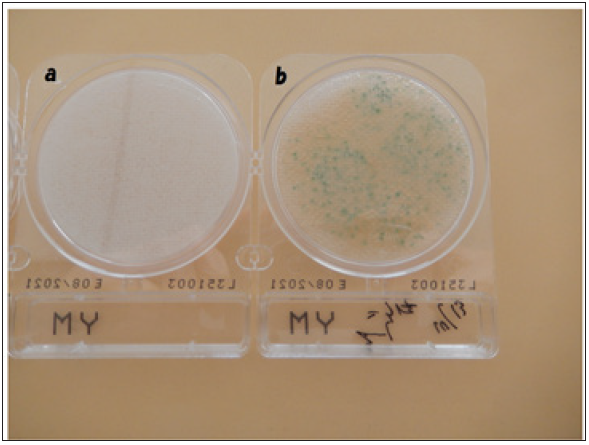
Figure 2: Microscopic observation of yeast in the malt from the antibiotic-producing co-culture used in Figure 1a.
Experiments at small scales and the antibiotic production after sub-culturing
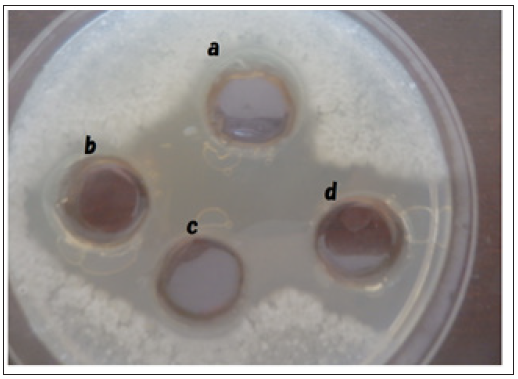
The next experiments were carried out using microfuge tubes containing 1-mL aliquots of malts. In short, DNA crown cells that were purified from 2mL of egg white were combined with yeast (approximately 3 mg/tube) in 1mL of D-MEM and incubated for 5 hours at 37 °C. Next, 50-µL aliquots of each co-culture were added to each of three tubes (designated a, b, and c) containing 1mL of malt each, and incubated for 5 days at room temperature. The resulting spent media were subjected to antibiotic testing. Clear zones were found for samples from all three tubes (data not shown), suggesting that antibiotic was produced at 5 days in these co-cultures. Next, 50-µL aliquots of the antibiotic-containing contents of the c tube were sub-cultured in each of 5 tubes (designated a-e) containing 1mL of fresh malt each. These sub-cultures were incubated for 5 days at 37 °C, and the resulting spent media again were subjected to antibiotic testing. Clear zones were found for samples from all five tubes (Figure 3), suggesting that antibiotic production persisted after sub-culturing.
Figure 3: Clear zones were observed in wells of samples from subculturing tubes a, b, c, and d. Initial co-cultures of DNA crown cells with yeast were carried out in 1mL malt for 5 days. After the detection of antibiotic in the malt, 50μL of the malt were added to each of 5 tubes of 1mL malt each and further incubated for another 5 days. Then, 400μL samples from each tube were subjected to antibiotic testing. Clear zones were observed for samples from all 5 tubes; the results for the first 4 tubes are shown in Figure 3.
On the other hand, no antibiotic production was observed in next round of sub-culturing. Regarding the mechanism of antibiotic production, many problems remain. One of these is that it takes a long time (5 weeks) until antibiotic is detected for co-cultures performed using egg white [8].To address this problem, DNA crown cells were purified from egg white and the resulting purified cells were used for the production of antibiotic. In this case, antibiotic production was found after 17 days of co-culturing of DNA crown cells and yeast [9], a result that was confirmed in the first experiment of the present study. Such results using cells without egg white suggested that the antibiotic production may be mediated by direct interaction of DNA crown cells and yeast. In the second experiment of the present study, DNA crown cells from 2mL of egg white were combined with yeast in small-scale (1-mL) cultures. In this case, antibiotic was produced after 5 days of co-culturing. At such a small scale (1mL), an increased ratio of contact is expected between DNA crown cells and yeast, and the concentration of antibiotics that are produced may be higher; as a result, antibiotic production may be detected at an earlier time (after 5 days) than was seen for co-cultures using malts at larger scales (e.g., volumes of 10L or 30mL).
On the other hand, to date, it remains unclear how contact between DNA crown cells and yeast is established in the co-cultures. It is well known that DNA from bacteria can be transferred into eukaryotes (trans-kingdom DNA transfer) [11], as has been seen under the microscope; however, it is very difficult to prove, by microscopic observation alone, whether antibiotic production in the present study involves the phenomenon of trans-kingdom DNA transfer [12]. In the present work, it was shown that yeast that had been co-cultured with DNA crown cells could not grow, an observation that correlated with antibiotic production. The results suggested that DNA crown cells may alter some aspects of yeast growth. It is well known that many genes, including those for antibiotic expression, are silent in yeast under normal conditions. We hypothesize that such genes may be expressed following stimulation by DNA crown cells.
As described above, it is possible that DNA crown cells cause some damage to yeast, possibly killing the yeast. In this case, it will be important to clarify how DNA crown cells stimulate or kill yeast. Such investigations also may clarify the function(s) of DNA crown cells.
Thus, based on the present and previous experiments, the following model is proposed for the mechanism of antibiotic production in systems employing the co-culture of DNA crown cells and yeast.
- The time until antibiotic is detected is influenced by the ratio of yeast and DNA crown cells and the
volume of malt used.
b. Antibiotic is produced as a result of direct contact between yeast cells and DNA crown cells.
c. Following the antibiotic production, the yeast die or otherwise become inviable. Any remaining yeast that continue to grow and produce antibiotics may remain in direct contact with DNA crown cells, which are providing some new functions that contribute to the maintenance or killing of these yeast.
Several problems remain to be addressed to confirm this proposed model. Notably, it is unclear whether DNA crown cells maintain these proposed functions for extended intervals (e.g., for 5 weeks in 10-L volumes of malt), how yeast are killed or inactivated, etc.
Acknowledgement
I would like to thank M. Hayakawa and H. Yamamura (Yamanashi University) for the supply of Streptomyces grises and for useful discussion. Also, I would like to thank I. Monna (Rizo Inc.) for assistance in extracting samples (DNA) and for useful discussions, S. Higuchi (Saitama Industrial Technology Center Northern Laboratory, Japan) for assistance with analysis of the TLC and for useful discussions, and Tokyo Metropolitan Research Industrial Technology Institute for the use of laboratory space and fluorescence microscopes.
References
- Inooka S (2012) Preparation and cultivation of artificial cells. App Cell Biol 25: 13-18.
- Inooka S (2016) Preparation of artificial cells using eggs with sphingosine-DNA. J Chem Eng Process Technol 7(1): 1-5.
- Inooka S (2016) Aggregation of sphingosine-DNA and cell construction using components from egg white. Integrative Molecular Medicine 3(6): 1-5.
- Inooka S (2017) Systematic preparation of bovine meat DNA crown cells. App Cell Biol Japan 30: 13-16.
- Inooka S (2018) Systematic preparation of DNA (Akoya pearl oyster) crown cells. App Cell Biol Japan 31: 21-34.
- Inooka S (2013) Preparation of artificial cells for yogurt production. App Cell Biol 26: 13-17.
- Inooka S (2014) Preparation of artificial human placental cells. App Cell Biol 27: 43-49.
- Inooka S (2019) Preparation of generated DNA(Streptomyces griseus) crown cells (artificial cells) and antibiotic production in its’ co-cultures with yeast (Beer). Current Trends on Biotechnology & Microbiology 1(2): 26-32.
- Inooka S (2019) Antibiotic production in co-cultures of DNA (Streptomyces) crown cells (artificial cells) and yeast (beer). Chem Pharm Res 1(2): 1-4.
- Inooka S (2017) Biotechnical and systematic preparation of artificial cells (DNA Crown Cells). Global Journal of Research in Engineering 17(1): 1-11.
- Katas BJ, Jablanovic J, Sun L, Ma L, Goldgof GM, et al. (2013) Direct transfer of whole genomes from bacteria to yeast. Nat Methods 10(5): 410-412.
- Husnik F, Mccutcheon JP (2018) Functional horizontal gene transfer from bacteria to eukaryote. Nat Rev Microbiol 16(2): 67-79.
© 2020 Shoshi Inooka. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






