- Submissions

Full Text
Novel Research in Sciences
Novel Anatomy and DNA Fingerprint of Deverra Tortuosa (Desf) DC, Syns: Pituranthos Tortuosus Benth
Assy NS1, Azazzy MF2* and El-Alfy TS3
1Faculty of Pharmacy, Egypt
2Department environmental studies and research institute, Egypt
3Professor of Pharmacognosy, Egypt
*Corresponding author:Azazzy MF, Department environmental studies and research institute, Egypt
Submission: May 22, 2019; Published: July 11, 2019
.jpg)
ISSN : 2688-836XVolume1 Issue3
Abstract
Deverra Tortuosa (Desf) DC is a perennial bushy desert medicinal and aromatic plant belongs to family Apiaceae, strongly aromatic shrub of 30-80cm height. The anatomy of the different organs of the plant has been demonstrated for the first time. The cross section of the leaf exhibited unusual type of structure characteristic to the plant. Transverse sections of the stem and root with special anatomy. DNA fingerprint has been made for the first-time using RAPD analysis, 9 primers with known sequences has been added, 6 of them has been positively reacted with mean value 2.33%. The present study demonstrates for the first-time additional tool to the taxonomical studies and the quality control testing for identification of Deverra tortuosa (Desf) DC in the field of pharmacogenetic studies and helps detect adulterations in the remedies of herbal medicines.
Introduction
A variety of herbal plants has been extensively used for the extraction of natural pure compounds used in the development of new and safe drugs. One of the most important therapeutic priorities in handling pain involves pain control; Apiaceae (Umbelliferae) is one of the important families in the flora of Egypt. The genus Deverra with two species (D. tortuosa and D. triradiata) these species are widespread in (Wadis) especially the tributaries of Wadi El Natron [1]. D. tortuosa is a perennial bushy; it grows naturally in sandy and stony plains [2]. The Devera sp. is known in traditional local medicine for the treatment of hypertension, against constipation, and in the case of bites [3]. The tender shoots and leaves are used as a condiment for the Bedouins local population. Pituranthos Tortuosus (Desf.) recently known Deverra Tortuosa (Desf.) DC is known in arabic as Shabat El-Gabal and is used in folk medicine as diuretic, carminative, analgesic, it is also used to relief stomach pain and against intestinal parasites [4]. The present study aims to demonstrate the anatomy and DNA fingerprint for Deverra Tortuosa (Desf) DC.
Materials and Methods
Fresh wild plants of Deverra Tortuosa (Desf) DC were collected from the Sadat city desert, Menofiya governorate at flowering stage on April 2014. The taxonomic identification of the plant material was confirmed using well identified herbarium specimens of Environmental studies and Research Institute and the Herbarium of Faculty of Science botany department, Mansoura University. A voucher specimen (ESRI Hr221) was deposited at the Herbarium of Environmental Studies and Research Institute (ESRI), University of Sadat city.
Anatomical study
The plant samples were stored by keeping them in 70% alcohol in the field studies conducted for the purpose of the anatomical structure studies of the plant parts. Samples were turned into permanent with glycerin-gelatin by taking cross-sections from their root, stem, flower and leaf [5]. The features of the sections, which were examined by light microscope, taking the photographs of the sections and transferring them to the digital media with Olympus microscope were actualized by using Spot in-sight color digital camera. The scales suitable for the lens size, of which the photographs are taken, were added to the images within the same program.
Powdered microscopy
Shade dried leaves, stem, fruit and roots were finely powdered and studied under microscope. Small quantity of different plant parts powder was placed separately on slides and each slide was mounted 2-3 drops of chloral hydrate and each slide was covered with cover slip then examined under microscope. Different cell components i.e. cork cells, sieve tubes fibers, lignified fibers, cortex cells, calcium oxalate crystals, mesocarp, endocarp and stomatal cells were noted, and photography was done by using digital camera [6].
DNA fingerprint study
Molecular study: Young leaves of Deverra Tortuosa (Desf) DC were collected and were grinded to a fine powder. Genomic DNA can be readily isolated from plant tissues of many species using the modification of the cetyltrimethylammonium bromide (CTAB) method reported by [7].
I. RAPD analysis:
a. The reaction proceeds in three steps: Heat denaturation of DNA double helix producing two single strands.
b. Annealing step in which the primers anneal, or attach, to the DNA template.
c. Elongation step during which the polymerase enzyme attaches strand-complementary nucleotides to the 3 end of the primer and continues to synthesize the complementary strand.
To obtain a satisfactory amount of DNA these three steps must be repeated in 20 or more cycles [8,9]. Random amplified polymorphic DNA (RAPD) was conducted according to [10].
II. Agarose gel electrophoresis:
Agarose gel electrophoresis was used to separate the fragments from the template DNA7.
a. The fragments were visualized by UV light using a transilluminator and were analyzed by gel Documentation system (Gene tool version 4, Syngene, Cambridge).
Statistical analysis
Data were expressed as mean±standard deviation. One-way analysis of variance (ANOVA) and Pearson correlation was used to analyze the obtained results with SPSS 17.0 software package. Difference on statistical analysis of data were considered significant at P<0.05.
Result & Discussion
Anatomy of Deverra Tortuosa (Desf) DC
The anatomy of the leaf: characterized with unusual type of anatomy of dicot leaf due to: the numerous vascular bundles which are the chief character of monocot leaves but the bundle with the same structure of dicot leaf, very reduced vascular bundles (reduced phloem and xylem) since the leaf itself is reduced to a scale leaf, presence of schizogenesis ducts on the top of each bundle to secrete and store volatile oil, formation of sunken stomata since the plant is a typical desert bush, presence of 2-3 rows of palisade tissue towards the lower epidermis, the lower epidermis of two layers with thicker cuticles than that of the upper epidermis which is made up of only one layer and thin cuticle (Figure 1 & 2). N.B. the lower epidermis is modified to take the role of the upper epidermis due the direct incidence of sun rays on its cells; the palisade tissue was formed underneath the lower epidermis and the sunken stomata were found for gas exchange during photosynthesis, and the thick cuticle was formed to reduce water loss due to the Xerophytic nature of this plant.
Anatomy of the root: Characterized by presence of periderm (cork tissue, cork cambium and phelloderm). Very reduced schizogenous ducts, presence of secondary tissue (xylem and phloem), the cambium ring is found due to the secondary thickening. The medullary rays divide the root section into two indicating two arms of xylem and phloem (Figure 7).
Anatomy of flower: Characterized by inferior ovary, two carpels and two locules, each locule with one ovule in axial placentation. One locule with mature ovule and the other locule with reduced ovule. Each locule is surrounded with six vitae, warty and hairy ovary wall, and flower epigynous. Presence of five stamens, stamen with two lobes each lobe with two pollen sacs full of pollen grains. Calyx with five reduced sepals, corolla with 5 petals. Presence of one schizogenous gland on the stigma (Figure 8 & 9)
Anatomy of the fruit: T.S. of the fruit characterized by schizocarpic fruit of two mericarps produces only one seed from the mature ovule (Figure 10-12). Each mericarp has six vitae. Warty and hairy wall of the fruit, generally, the plants of this family characterized by the presence of schizogenous glands in all organs (leaf, stem, root, flower and fruit) [11], warty and hairy ovary wall [12]. Pharmacogenetic and physicochemical studies, being reliable and inexpensive, play an important role in quality control issues of the crude drug samples [13]. Morphological and microscopical studies of stem, leaf and root will be helpful in the identification of these parts of Deverra Tortuosa (Desf) DC plant. In pharmacogenetic studies different types of evaluations were carried out that focus on microscopical examination [14]. The oil glands (schizogenous ducts) are found in large sizes and numbers in both leaf and stem and in flowers and fruits vittae are found from five to six around each locule. However, the oil glands (schizogenous ducts) are observed in the root but in small number and very reduced. Our observations in this study through thin layer chromatography (TLC) recorded the absence of essential oil in the root, which may explain the much reduction in number and size of oil glands (schizogenous ducts) in the root. The presence of the essential oil in the other organs such as flower, stem and leaf may explain the abundance of oil glands in large sizes and numbers where the oil is excreted and stored.
Figure 1:Detailed T.S. of Deverra Tortuosa leaf. s.st: Sunkin stomata t.cu.: Thick cuticle, l.ep: Lower epidermis pal.t: palisade tissue u.ep: Upper epidermis par.c: Parenchyma cells g.c: guard cell x.v: xylem vessels ph: phloem v.b: vascular bundle sch.d: schizogenous duct tn.cu: Thin cuticle
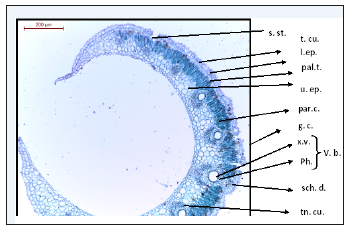
Figure 2:T.S. of Deverra Tortuosa leaf showing a comparison of lower and upper epidermis.
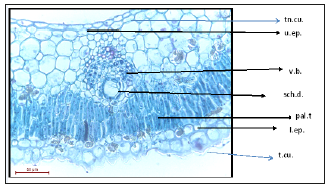
Figure 3:T.S. of Deverra Tortuosa stem. w.c: warty cuticle s.s: sunkin stomata e.p: epidermis chlo: chlorenchyma palisade like tissue (2-3 rows) a.col: Angular collenchyma sch.d: schizogenous duct ph: phloem, cam: Cambium m.x: metaxylem p.x: protoxylem a.par: Angular parenchyma (pith)
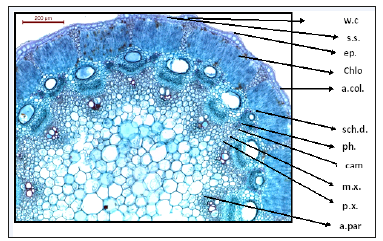
Figure 4:T.S of Deverra Tortuosa stem showing two layers of epidermis and five rows of chlorenchyma palisade like tissue.
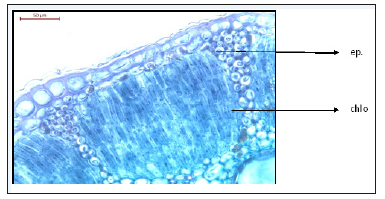
Figure 5:T.S. of Deverra Tortuosa stem showing large schizogenous ducts.
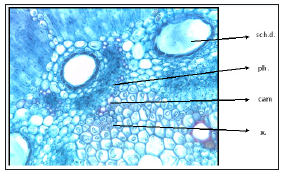
Figure 6:T.S. of Deverra Tortuosa stem showing metaxylem and protoxylem.

Figure 7:T.S. of Deverra Tortuosa root. r. sch.d: reduced schizogenous duct c.c.: cork cells c.cam: cork cambium phil: philloderm p: periderm s.ph: Secondary phloem m.r: medullary ray p.x: primary xylem s.x: secondary xylem cam. r: cambium ring
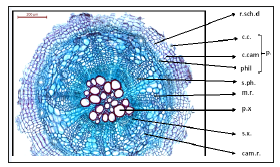
Figure 8:T.S. of Deverra Tortuosa flower. co: Corolla stg: stigma an: Anther sch.d. schizogenous duct p.sac: pollen sac ca: calyx st.: style r.o: Reduced ovule o: ovary m.o: mature ovule v: vitae w & h.s: warty and hairy surface
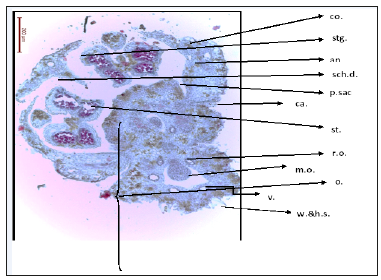
Figure 9:T.S. of Deverra Tortuosa flower showing large schizogenous duct in the middle of the stigma.
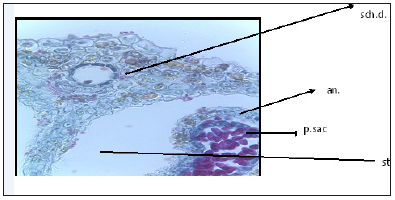
Figure 10:T.S. of Deverra Tortuosa fruit. w. & h. w. warty and hairy wall sch.d: schizogenous duct o.s: one seed r.o: reduced ovule
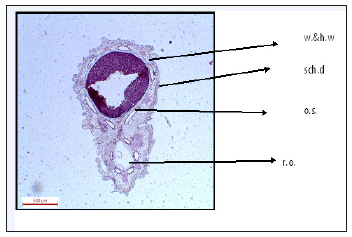
Figure 11:The main characteristic elements of Deverra Tortuosa plant powder.

Figure 12:RAPD-PCR amplification fragment generated by 9 primers of DNA extracted from Deverra Tortuosa (Desf) DC leaf.
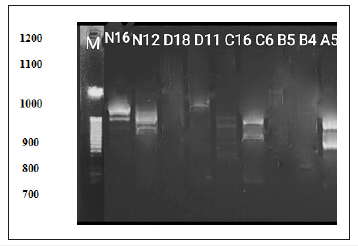
DNA fingerprint: Molecular marker system has a significant advanced understanding of plant genomes; RAPD is successfully employed for varied purposes in plant genetics and breeding [15]. The usefulness of RAPD for studying the genetic relationships and identifying taxa within a genus has been proved [16]. Data obtained showed in (Table 1 & Figure 1) revealed molecular identification of Deverra Tortuosa was carried out using 9 RAPD-PCR primers. The total of 14 bands was amplified.
Table 1:Type of primers used to amplify DNA extracted from Deverra Tortuosa (Desf) DC; the total number of RAPDPCR and size of fragments by base pair.
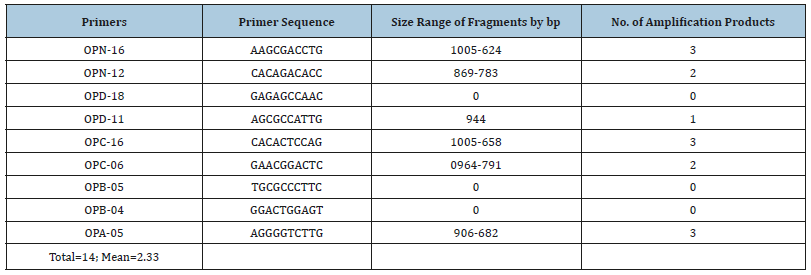
DNA profile amplified by RAPD-PCR: The Random amplified polymorphic DNA was used to generate fingerprint pattern for Deverra Tortuosa (Desf) DC grown wildly in Sadat desert area (Western Desert). From a total of 15 oligonucleotide primers only six generated amplified fragments. A total of 14 marker fragments were amplified with mean 2.33. Gel electrophoresis indicated a variable size of fragments ranged from 1005 to 624 base pair. Primer OPN-16 generated 3 reproducible fragments ranged from 1005 to 624bp; OPC-16 primer generated 3 reproducible fragments ranged from 1005 to 658bp; while OPA- 05 generated 3 fragments ranged from 906 to 682bp. The least number of bands were produced by OPD-11 primer generating only one reproducible fragment with molecular size 944bp. The advantage of molecular markers over phenotypic data is the possibility to compare genotypes, even if they are sampled in different environment, type of tissue or stage of development. Another advantage is theoretical possibility to detect poly morphism through the entire genome [17]. These results of the present study indicated that the molecular markers that can be used in the identification of Pituranthos Tortuosus and differentiation from other related species. The RAPD markers cover large parts of genomic DNA a and provide variable information to estimate genetic variability [18,19]. Molecular markers play an important role in many aspects of plant breeding, such as identification of the genes responsible for desirable traits. Molecular markers have been widely used to map important genes and assist with the breeding of oil crops [20].
Molecular, genomic and molecular breeding research on Deverra Tortuosa has been restricted due to the limited availability of genomic and transcriptomic sequence data. High-throughput RNA-seq can generate massive amounts of data on whole-genomewide transcripts and has become more cost efficient [21]. In the present study, the fingerprint of Deverra type was identified. These unigenes can serve as a reference base for the future identification of trait-associated single-nucleotide polymorphism and SSR markers that can be used in marker-assisted in breeding programs. The data can also serve as a resource for gene expression analyses, and the identification and characterization of celery genes with horticultural importance [22]. The analysis of the amplified fragments generated by RAPD reactions revealed that the genetic profile of Deverra Tortuosa produced diverse molecular patterns. The primers OPC-16 (Primer Sequence CACACTCCAG Size range of fragments by 1005-658), while, OPN-16 (Primer Sequence AAGCGACCTG and Size range of fragments by bp 1005-624) can be used for the identification of this species since they generate producible fragments. These results indicated the molecular markers that can be used in the identification of Deverra Tortuosa (Desf) DC and differentiation from other related species. Such DNA fingerprint study is useful in case of substituted or adulterated plants with other species or varieties that are morphologically indistinguishable also can resolves adulteration problems faced by majority of the herbalists, pharmaceutical industries and practitioners [23].
Acknowledgment
The authors are sincerely grateful to Professor Sekina Mohamed Kamel Ayyad Professor of Botany, Faculty of Science, Mansura University for continuous supporting this work. All thanks are due to Professor Sahar Abd El-Tawaab Professor of Genetics; Faculty of Ain Shams for girls, for housing the DNA experiment in her laboratory, her commenting on the results is very much appreciated. Also, for Professor Magda Soliman, Professor of Genetics, Faculty of Science, Mansoura University, for revising the manuscript. Thanks also due to Professor Refaey M Professor of Geology, Surveys of Natural Resources Department. Environmental Studies and Research Institute, University of Sadat City for his encouragement and support.
References
- Azzazi MF (2011) Morphological studies of the pollen grains of wadi El- Natrun plants, West Nile delta, Egypt. Plant Syst Evol 294(3-4): 239-251.
- Boulos L (2000) Flora of Egypt. AL-Hadara-Publishing, Cairo, Egypt, 2.
- El-Mokasabi FM (2014) Floristic composition and traditional uses of plant species at Wadi Alkuf, Al- Jabal Al-Akhder, Libya. American- Eurasian J Agric Environ Sci 14(8): 685-697.
- Mahran GH, Ahmed MS, Seida AA, Amarquaye AA (1989) Bulletin of Faculty of Pharmacy, Cairo University, Egypt, 27(1): 87.
- Vardar Y (1987) Botanikte preparasyon teknigi. Ege University Science Faculty Press, Izmir, Turkey, pp. 1-66.
- Mehlhorn H (2011) Nature helps: How plants and other organisms contribute to solve health problems. Springer, Germany, p. 12.
- Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19: 11-15.
- Wilson K, Walker J (2000) Principles and techniques of practical biochemistry. Cambridge University Press, United Kingdom, p. 784.
- Reece W (2004) Criminology and genetically modified food. British Journal of Criminology 44(2): 151-167.
- Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18(22): 6531-6535.
- Akpulat HA, Ataşlar E (2015) The anatomical and micromorphological structure of peucedanum ozhatayiorum (2010).
- Pedro, JM, Pablo V (2015) Taxonomy of the tribe Apieae (Apiaceae) revisited as revealed by molecular phylogenies and morphological characters. Phytotaxa 212(1): 57-79.
- Bigoniya P, Singh CS, Srivastava B (2012) Pharmacogenetic and Physicochemical standardization of Syzigium cumini and Azadirachta indica seed. Asian Pac J Trop Biomed: S290e5.
- Evans WC (2009) Trease and Evans pharmacognosy. (16th edn), Saunders Ltd., Philadelphia, USA, p. 108.
- Vala AK, Shah S, Patel R (2014) Biogenesis of silver nanoparticles by marine derived fungus Aspergillus flavus from Bhavnagar coast, gulf of Khambhat, India. J Mar Biol Oceanogr 3(1): 1-3.
- Soliman MI, Rizk RR, Mohammed NS (2014) Genetic characterization and relationships among Egyptian cultivated and wild Trifolium L. as revealed by karyological, biochemical and molecular markers. Journal of Biological Chemistry and Environmental Sciences 9(2): 219-243.
- Sakiyama NS, Ramos HCC, Caixeta ET, Pereira MG (2014) Plant breeding with marker-assisted selection in Brazil. Crop Breed Appl Biotechnol 14(1): 54-60.
- Singh N, Pal AK, Roy RK, Tewari SK, Tamta S, et al. (2016) Assessment of genetic variation and population structure in Indian Gladiolus cultivars inferred from molecular markers. The Nucleus 59(3): 235-244.
- Thorat AS, Sonone NA, Choudhari VV, Devarumath RM, Babu KH (2017) Plant regeneration from cell suspension culture in Saccharum officinarum L. and ascertaining of genetic fidelity through RAPD and ISSR markers. 3 Biotech 7(1): 16.
- Wenliang W, Xiaoqiong Q, Linhai W, Yanxin Z, Wei H, et al. (2011) Characterization of the sesame (Sesamum indicum L.) global transcriptome using Illumina paired-end sequencing and development of EST-SSR markers. BMC Genomics 12: 451.
- Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: A revolutionary tool for transcriptomics. Nature Reviews Genetics 10(1): 57-63.
- Nicolai M, Pisani C, Bouchet JP, Vuylsteke M, Palloix A (2012) Discovery of a large set of SNP and SSR genetic markers by high-throughput sequencing of pepper (Capsicum annuum). Genet Mol Res 11(3): 2295- 2300.
- Srivastava S, Mishra N (2009) Genetic Markers a cutting-edge technology in herbal drug research. Journal of Chemical and Pharmaceutical Research 1(1): 1-18.
© 2019 Azazzy MF . This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






