- Submissions

Full Text
Novel Approaches in Cancer Study
A Prostate Cancer Specific Telomerase Repressor Region on Human Chromosome 11q
Rana Hasan1, Hemad Yasaei2, Sheila Matta1, Alison Marriott1, Robert Newbold1 and Terry Roberts1*
1Department of Health and Life Sciences, Department Biosciences, Brunel University London, Uxbridge, Middlesex, UK
2Dubai Genetics Centre, Dubai Health Authority, United Arab Emirates
*Corresponding author:Terry Roberts, College of Health and Life Sciences, Department of Life Sciences, Biosciences, Brunel University London, Uxbridge, Middlesex, UB8 3PH, UK
Submission: October 21, 2022 Published: October 31, 2022

ISSN:2637-773XVolume7 Issue3
Abstract
Telomerase reactivation is one of the key events in the development of cancer. More than 90% of all cancers de-repress sequences within the genome, which results in the overexpression of hTERT and telomerase. In doing so, telomeres at the ends of human chromosomes, which would naturally shorten after each round of replication, are maintained indefinitely giving rise to immortalized cells. The actual mechanisms responsible for telomerase depression in cancer are still under intense investigation and the elucidation of this process would represent a major therapeutic target. The literature has pointed to genes on different chromosomes being responsible for telomerase repression in specific cancers. Microcell Mediated Chromosome Transfer (MMCT) was used to transfer human chromosomes 3, 8, 10, 11, 13, and 17 into the prostate cancer cell line PC3. In doing so, we observed robust and sustained hTERT and telomerase repression for chromosome 11. We then fragmented chromosome 11 using 25G of gamma radiation before repeating the transfer into PC3. We found that 70% of the hybrid clones picked expressed very low levels of hTERT and telomerase when compared to the controls. STS mapping of the clones identified 3 distinct regions on chromosome 11q which could harbour sequences responsible for telomerase repression specific to prostate cancer. Identification of this telomerase repressor could lead to new chemotherapeutic drugs to target prostate cancer or a highly specific biomarker for the disease.
Keywords:Prostate cancer; Telomerase; hTERT; MMCT; Chromosome 11; Mapping
Introduction
More than 90% of all known cancers reactivate the enzyme telomerase as part of the pathway leading to cellular immortalization, transformation and eventually tumorigenesis [1]. The telomerase enzyme itself is composed of a catalytic protein subunit coded for by the hTERT gene and an RNA (TR) subunit [2]. Telomerase regulation has been extensively studied in different biological systems, this has uncovered several mechanisms and genes which activates telomerase expression in cancers. hTERT core promoter mutations that generate novel ETS transcription factor binding sites (-124C>T and -146C>T in relation to the transcriptional start site), have been identified in several tumour types [3,4] These mutations cause moderate activation of the hTERT promoter which allows cells to delay replicative senescence but not enough for full activation and thereby immortality. Interestingly, two independent studies [5,6] concluded that mutations within the core promoter region of hTERT is a rare event in prostate cancer, both groups did not identify mutations in the samples that were analysed. Recently, CpG dinucleotide hypermethylation within the THOR (TERT Hypermethylated Oncological Region) region of the hTERT promoter (-649 to -217) has been shown to activate hTERT expression [7,8]. In cancers that exhibit high telomerase expression, the THOR region remains heavily methylated whereas normal cells contain significantly less methylation in this region. Furthermore, the promoter region of hTERT is subject to regulation by several well-known transcriptional activators and repressors [9]. The oncogene c-MYC directly activates hTERT through binding to the e-Box region of the core promoter [10] and overexpression of c-MYC in prostate cancer is linked to disease progression [11] suggesting that c-MYC could be a potential mechanism responsible for hTERT depression in prostate cancers. Cuthbert et al [12] identified a region on human chromosome 3p that was responsible for hTERT repression in the HER2 positive breast cancer cell line 21NT. Later in 2003, [13] found that a sequence located on human chromosome 3 regulated telomerase expression by acting directly within Intron-2 of the hTERT gene on chromosome 5. In addition to the mechanisms described above, the regulation of telomerase activity was found to be controlled by the rate of transcription of the hTERT gene [14], and that intron-2 containing pre-spliced hTERT mRNA correlates with telomerase activity. Studies on early-stage prostate cancer using CGH have shown that the loss of genetic material was higher than the gain, which suggests that inactivation and loss of tumour suppressor genes are involved in prostate cancer predisposition [15]. The loss of genetic material has been reported to occur most frequently on chromosomes 2q, 5q, 6q, 8p, 10q, 13q, 16q and 18q. Chromosomal regions 8p and 13q were shown to be lost in over 50% of high-grade PIN (Prostatic Intraepithelial Neoplasia). This strongly suggests that these regions harbour genes which are involved in prostate cancer tumorigenesis. Chromosomal regions 2p, 11p, 1q, 3q, 4q, 7q, 8q, 11q, 12q and Xq been reported to show gains in genetic material which could involve the activation of oncogenes [16-18]. Recent work has shown that up to 100% of prostate tumours show the reactivation of telomerase and is therefore a common feature in this type of cancer [19,20]. Several groups have now linked chromosome 11 to prostate cancer development and the 11q region is under intense investigation [21-23]. Zheng et al. Chung et al. and 2012 linked prostate cancer risk and predisposition to 11q13 whereas [24] carried out fine mapping using SNP’s and identified a region at 11q13.5 which was associated with prostate cancer deaths and identified DGAT2 at 11q14.5 as a potential gene involved in prostate cancer risk [25]. We have previously successfully identified and mapped the breast cancer telomerase repressor region to human chromosome 3p21- 22 [12-14], and we have applied the same principles to identify chromosomes involved in repression of telomerase in prostate cancer.
Result
Identification of a human chromosome which represses hTERT
By using the same methodology as Cuthbert et al. [12], we wanted to identify a human chromosome that could repress hTERT expression and hence telomerase in the prostate cancer cell line PC3, as chromosome 3 did in the breast cancer cell line 21NT. Previous publications by our group [12-14] have shown that transfer of human chromosome 3 into the breast cancer cell line 21NT resulted in the downregulation of hTERT/telomerase leading to cellular senescence within 4 weeks. This left us with a small window of time in which to collect sufficient material to perform downstream molecular analysis as the cells were growing slowly. We therefore assume that the same scenario would occur in the PC3 cell line. To prevent this from occurring in this work, we cloned the full-length hTERT cDNA into the plasmid vector PCl-neo (Promega, UK) and transfected it into PC3 cell. Resulting clones stably expressed exogenous hTERT using the CMV promoter were picked and designated PC3/hTERT. These PC3/hTERT cells do not enter senescence when an intact human chromosome containing telomerase repressive sequences is transferred into them due to the high levels of exogenous hTERT expression. This allowed cells to grow at a normal rate and more importantly, it allowed us to quantify the endogenously transcribed pre-spliced intron 2 containing hTERT mRNA.
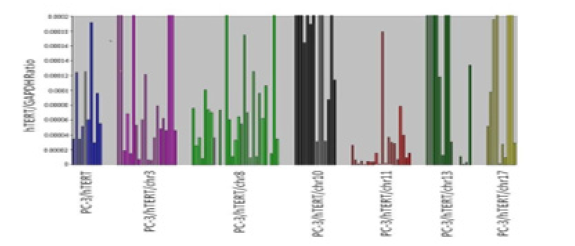
We transferred whole human chromosomes 3, 8, 10, 11, 13 and 17 into the PC3/hTERT cell line. Endogenous hTERT expression analysis using qPCR was then performed to determine which chromosomes repressed hTERT. We found that most of the chromosome 11 hybrids (18/20) expressed 50% lower level of hTERT than the parental host cell line PC3/hTERT. In other words, 9 PC3/hTERT clones expressed 8.33 x 10-5 hTERT transcripts, whereas 18 chromosome 11 hybrids expressed less than 4.16 x x10-5 transcripts. (Figure 1). Furthermore, 50% of the chromosome 11 hybrids (10/20) expressed less hTERT transcripts than 10% of the average PC3/hTERT clones. In comparison, the repression of hTERT seen in the chromosome 11 hybrids was much stronger than that seen in PC3/hTERT chromosomal hybrids 3, 8, 10, 11, 13, and 17. An unpaired t-test between hTERT expression levels in all chromosomal hybrids against the PC3/hTERT control revealed a statistically significant difference of p=0.0039 for the chromosomal 11 hybrids.
Figure 1: Comparison of endogenous hTERT mRNA levels in individual hybrid clones generated by transferring normal human chromosomes 3, 8, 19, 11, 13 and 17 into PC3/hTERT cells. GAPDH was used as the endogenous control and all qPCR reactions were done in triplicate for both the target and endogenous control gene.
Repression of pre-spliced hTERT by human chromosome 11
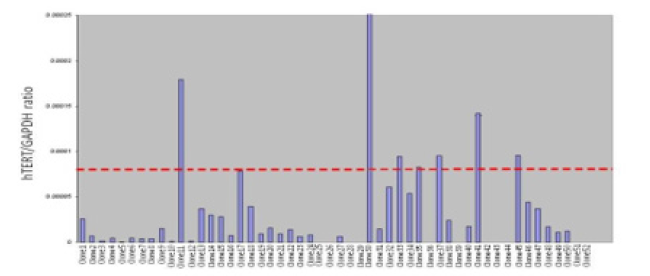
Human chromosome 11 was transferred into the PC3/hTERT cell line using MMCT, this was repeated 3 times and a total of 52 clones were picked and analysed for hTERT expression using qPCR to confirm robust levels hTERT repression (Figure 2). The mean levels of hTERT expression in the control cells, PC3/hTERT, was compared with the hTERT levels in the PC3/hTERT chromosome 11 hybrids. We found that 21% (11/52) of the hybrids showed no detectable expression of hTERT which suggests complete repression of the gene. Furthermore, when the average expression level of the control cell line PC3/hTERT (shown as a red dotted line in Figure 2) was compared with the PC3/hTERT chromosome 11 hybrids, 47% (24/52) of the hybrids had lower hTERT expression levels than 12.5% of the controls. Interestingly, 79% (41/52) of the chromosomal hybrids displayed hTERT expression levels lower than 50% of the control mean.
Figure 2: Repeat transfer of normal human chromosome 11 into PC3/hTERT cells using MMCT. 52 individual hybrids were picked and hTERT analysis performed. The red dotted line indicates the average hTERT expression level in the control cell line PC3/hTERT.
TRAP analysis of hybrids containing whole human chromosome 11
To confirm the result, we obtained from the qPCR in Figures 1 & 2, we repeated the human chromosome 11 transfer into PC3 cells and 17 individual hybrids were picked. TRAP analysis was then performed on the hybrid clones with PC3 being used as the control. From Figure 3a, 3b, it can be seen that 15 hybrids containing human chromosome 11 show robust repression of telomerase activity when compared to the PC3 control.
Figure 3: Telomerase activity in 17 hybrid clones generated by the transfer of normal human chromosome 11 into the PC3 cell line. (a) Shows a representative TRAP gel containing TRAP ladders, Image J was used to take densitometry readings of the TRAP bands and the values plotted on the histogram. (b) Represents the corresponding histogram showing the levels of telomerase activity in the hybrid clones compared to the parental PC3 cell line. The positive control is shown in lane 1 and lane 2 shows the heat inactivated negative control.

Fine mapping of the prostate telomerase repressor to human chromosome 11
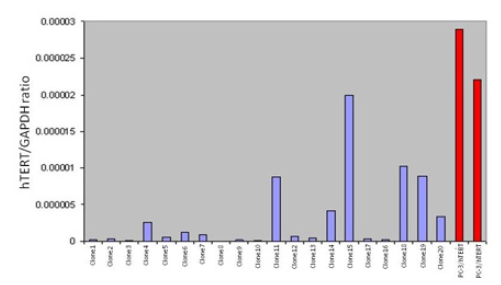
Exposure of microcells to 25 Grays of radiation successfully fragmented human chromosome 11. The smaller fragments were then transferred into PC3/hTERT to produce a mixture of chromosome 11 hybrid clones that are hTERT repressed or nonrepressed. The repression / non-repression of hTERT would depend on the chromosome 11 fragment that is picked up by the hybrids. 70% of the hybrids (14/20) contained very low levels of endogenous hTERT mRNA compared with the control PC3/hTERT cell line (Figure 4). In addition, 13/20 clones (65%) had almost completely silenced hTERT transcription, strongly suggesting that the telomerase / hTERT repressor gene was present on these fragments. We hypothesise that the 13 hybrids, which repress hTERT share a common region of chromosome 11 and that the same repressive element was responsible for the result obtained.
Figure 4: XMMCT (irradiated microcell mediated chromosome transfer). Microcells containing whole chromosome 11 was fragmented using 25G gamma irradiation prior to transfer into PC3/hTERT cells. 20 individual hybrids containing small fragments of normal human chromosome 11 were picked and endogenous hTERT expression quantified and compared to the parental PC3/hTERT cell line.
Chromosome paint for fragmented chromosome 11 hybrid clone 1
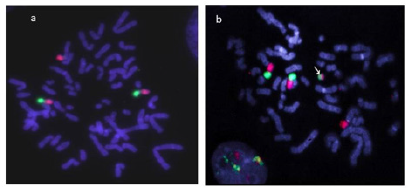
To examine the integrity of the transferred chromosome 11 fragment, we performed Fluorescence In-Situ Hybridization (FISH) with chromosome 11 arm specific probes. This analysis would show us the number of chromosome 11 fragments present in the hybrid and if the fragment is a discrete “mini-chromosome” or if it has translocated onto a host chromosome. Hybrid clones containing fragmented chromosome 11 and display repressed hTERT levels were used to prepare metaphase spreads. The spreads were then analysed using p-arm specific probes labelled with FITC, and q-arm specific probes labelled with Texas Red. All the strongly repressed (hTERT mRNA) hybrids contained a single discrete chromosome 11 fragment which had both p and q arm chromosomal DNA (representative image in Figure 5). The chromosome 11 fragments appeared as a “mini-chromosome” in all the hTERT repressed hybrids that were analysed by FISH.
Figure 5: FISH analysis of PC3/hTERT hybrid clone 1 (from Figure 4) using chromosome 11 arm specific paints (green= p-arm, red= q-arm). The recipient cell line (a) shows the presence of two chromosome 11 plus an additional copy of 11q translocated onto another chromosome. In (b) which represents PC3/hTERT hybrid clone 1, the same pattern of chromosome 11 is seen with the addition of a new “mini” chromosome (arrow). This is the fragment of normal chromosome 11 that was transferred into the recipient cells. This mini chromosome was then transferred back into a mouse A9 background for STS mapping.
STS mapping of hTERT repressing clone 1 from Figure 4
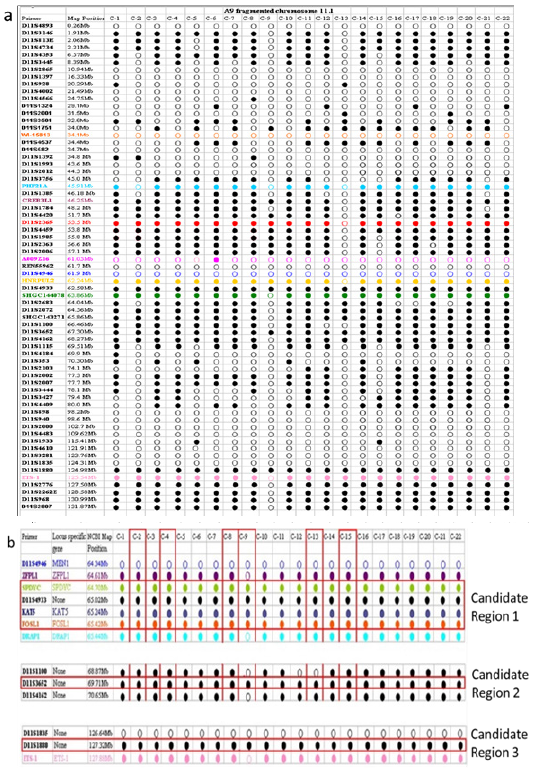
To further define the location of the prostate cancer specific hTERT repressor, we need to accurately map the fragments present in the radiation reduced chromosome 11 hybrids. The fragmentation of chromosome 11 should allow us to pinpoint specific regions on the chromosome by excluding regions not present in the transferred fragment. To map the chromosome 11 material present in hybrid clone 1 (PC3/hTERT/fchr11.1), we transferred the “mini-chromosome” back into a mouse A9 background using MMCT, picked 22 human: A9 hybrids and mapped them using human specific chromosome 11 STS (sequence tagged site) markers (NCBI). The STS map Figure 6a showed the presence of telomeres, p and q-arms, and a centromere in nearly all clones (except A9/fchr11.1.13 and A9/fchr11.1.9). From Figure 6a & 6b, three regions of commonality can be seen, we predict that these regions should harbour the prostate cancer specific telomerase repressor gene.
Figure 6:The Mini chromosome in hybrid clone 1 was transferred back into the mouse A9 background for fine mapping using human STS markers representing the full length of chromosome 11. In total, 22 clones were picked and mapped (a). To our surprise, not all the clones were identical, some had acquired additional deletions during the transfer. Based on the mapping information, the predicted sizes of the retained fragments range from 6Mb to 40Mb. Fine mapping of 3 candidate regions (b) which should harbour the telomerase repressor specific to prostate cancer.
Discussion
The synergistic effects of gamma radiation exposure of the microcells followed by the MMCT from PC3 cells back into the A9 background resulted in a complex series of fragments as seen in Figure 6a. This was fortuitous and advantageous as it allowed us to narrow down the region carrying our telomerase/hTERT repressor gene. From closer analysis of the hTERT repressive clones (Figure 6a), it is possible to narrow down the region, which should contain the prostate cancer telomerase repressor. This would be regions that are common to all the STS mapped hybrids. From our analysis, we predict that the repressor will lie within one of 3 regions of human chromosome 11q, this being the 64.7-65.4Mb, 69Mb and 127.3Mb regions (Figure 6b). This report is the first of its kind to identify a potential telomerase repressor region on human chromosome 11. Through the use of the MMCT technique, we have identified 3 region of human chromosome 11q that harbour potential telomerase repressor genes. To our knowledge, this is the first report of human chromosome 11 being involved in the regulation of telomerase. Identification of such genes would represent a major milestone for the development of drugs or biomarkers to specifically target prostate cancer. The regions we have identified on chromosome 11q (11q12-13) that harbour the prostate cancer telomerase repressor, coincides with regions previously predicted (11q13-11q14) to contain genes involved in prostate cancer progression [21-25]. Chromatin remodelling has been shown to be a major mechanism that regulates genes in mammalian cells [13]. Szutorisz et al. [13] identified two nuclease 1 sensitive sites within intron 2 of the hTERT gene that can be transcriptionally repressed through chromosome transfer. These nuclease -sensitive sites were shown to under-go chromatin remodelling to transcriptionally repress hTERT in cancer cells after transfer of a normal copy of human chromosome 3 [26]. We therefore predict that the telomerase repressor sequences could have chromatin remodelling activity. Based on this assumption, several genes within our candidate region fits this criterion. These are, BRMS1, a Histone Deacetylase (HDAC), KDM2A, a histone demethylase and KAT5, a Histone Acetyltransferase (HAT). These all map within the 61.08Mb-65.42Mb region of Figure 6. This will be the focus of our future investigations. Schleutker et al. [27] identified regions on human chromosome 11(11q14) and 3 (3p25) which contains genes predisposing to hereditary prostate cancer. We previously identified chromosomal region 3p22 as containing the breast cancer specific telomerase repressor 12-14] and in this report, we identified 11q12-13 as the prostate specific telomerase repressor region. It is known that breast and prostate cancer predisposition is linked and a history of one can lead to the risk of the other [28-30]. Therefore, telomerase regulation in these two cancers may also be linked. This will be investigated further during our search for this cancer specific telomerase regulating genes.
Materials and Method
Cell culture conditions
The epithelial prostate cancer cell line PC3, was cultured in Ham’s F-12 Nutrient mix (Fisher Scientific, UK) with 7%FBS and 2mM L-glutamine. Cells were grown at 37 ℃ in a fully humidified incubator and 6.5% CO2.
Metaphase preparation
Cells were grown to 70% confluence then treated with Demecolcine (0.1μg/ml) and incubated at 37 ℃ for 2-4 hours. Adherent cells were then trypsinised and pelleted by centrifugation at 1000 RPM for 5 minutes. Cells were then treated with prewarmed 0.075M KCl for 30 minutes, then fixed with methanol: acetic acid (3:1) solution, and washes 3 times with fixative solution. Cell pellets were resuspended in 1ml of fresh fixative and 20μl of the cell suspension dropped onto clean slides.
Fluorescence in Situ Hybridization (FISH)
q-arm specific chromosome 11 paint probes labelled with Texas Red and p-arm specific chromosome 11 paint probes labelled with FITC were obtained from XCAP-Meta Systems (Germany). FISH was carried out as mentioned in Linne et al. [26].
Micro-Cell Mediated Chromosome Transfer (MMCT)
The mono chromosomal hybrid panel (human: rodent) was developed and constructed by Professor R. Newbold’s group for gene identification, mapping and functional analysis. Whole, intact, individual human chromosomes and fragmented human chromosomes 11 were transferred into the PC3 prostate cancer cell line using the MMCT procedure as outlined previously [12,26].
RNA extraction and qPCR
RNA extraction for tissue culture dishes of 80% confluence was performed as outlined in Motevalli et al. [31]. qPCR for pre-spliced hTERT and GAPDH quantification using an absolute assay and Sequence Tagged Site microsatellite mapping (STS mapping) was performed as mentioned in [12,26].
Determination of telomerase activity using the TRAP assay
Individual hybrids containing either whole chromosome of chromosomal fragments was picked and grown in p60 dishes to a confluence of 80% as described above. The dishes were then trypsinised, the cells counted and 3-5x105 cells were pipetted into a chilled microfuge tube and spun down at 6000rpm for 3 minutes at 4 ℃. The supernatant was removed, the resulting pellet stored at -80 ℃ until needed. For analysis, the samples were thawed on wet ice and 200ul of cold 1x CHAPS lysis buffer (3-[(3-cholamidopropyl) di methyl ammonia]-1-propanesulfonate, Sigma-Aldrich, UK) was added to pellet, the sample was mixed by pipetting and then incubated on ice for 30 minutes. After incubation, the samples were centrifuged at 12000rpm for 20 minutes at 4 ℃. Approximately 160ul of the resulting aqueous supernatant (containing active telomerase enzyme) was removed to a fresh cold microfuge tube and place at -800C until needed. The Bradford protein assay (Coomassie Blue G-250, Thermofisher, UK) was performed on the supernatant to measure the protein content, and telomerase activity was determined using the TRAPeze® telomerase detection kit (Sigma-Aldrich, UK). TRAP gels were then scanned using the Bio-Rad ChemiDoc XRS system and images were scanned w image J. Densitometry readings of the TRAP bands obtained and used to plot a histogram of activity.
Acknowledgment
We would like to thank Brunel Universities College of Health, Life and medical Sciences for their support.
References
- Newbold RF (2002) The significance of telomerase activation and cellular immortalization in human cancer. Mutagenesis 17(6): 539-550.
- Wyatt HD, West SC, Beattie TL (2010) In TERT prating telomerase structure and function. Nucleic Acids Res 38(17): 5609-5622.
- Bell RJ, Rube HT, Xavier-Magalhaes A, Costa BM, Mancini A, et al. (2016) Understanding TERT Promoter Mutations: A Common Path to Immortality. Mol Cancer Res 14(4): 315-323.
- Rachakonda S, Hoheisel JD, Kumar R (2021) Occurrence, functionality and abundance of the TERT promoter mutations. Int J Cancer 149: 1852-1862.
- Stoehr R, Taubert H, Zinnall U, Giedl, J, Gaisa NT, et al. (2015) Frequency of TERT promoter mutations in prostate cancer. Pathobiology 82(2): 53-57.
- Wu S, Huang P, Li C, Huang Y, Li X (2014) Telomerase reverse transcriptase gene promoter mutations help discern the origin of urogenital tumors: a genomic and molecular study. Eur Urol 65(2): 274-277.
- Leao R, Apolonio JD, Lee D, Figueiredo A, Tabori U, et al. (2018) Mechanisms of human telomerase reverse transcriptase (hTERT) regulation: clinical impacts in cancer. J Biomed Sci 25(1): 22.
- Lee DD, Leao R, Komosa M, Gallo M, Zhang CH, et al. (2019) DNA hypermethylation within TERT promoter upregulates TERT expression in cancer. J Clin Invest 129(1): 223-229.
- Dratwa M, Wysoczanska B, Lacina P, Kubik T, Bogunia-Kubik K (2020) TERT-regulation and roles in cancer formation. Front Immunol 11: 589929.
- Wu KJ, Grandori C, Amacker M, Simon-Vermot N, Polack A, et al. (1999) Direct activation of TERT transcription by c-MYC. Nat Genet 21(2): 220-224.
- Hawksworth D, Ravindranath L, Chen Y, Furusato B, Sesterhenn IA, et al. (2010) Overexpression of C-MYC oncogene in prostate cancer predicts biochemical recurrence. Prostate Cancer Prostatic Dis 13(4): 311-315.
- Cuthbert AP, Bond J, Trott DA, Gill S, Broni J, et al. (1999) Telomerase repressor sequences on chromosome 3 and induction of permanent growth arrest in human breast cancer cells. J Natl Cancer Inst 9(1): 37-45.
- Szutorisz H, Lingner J, Cuthbert AP, Trott DA, Newbold RF, et al. (2003) A Chromosome 3-encoded repressor of the human Telomerase Reverse Transcriptase (hTERT) gene controls the state of hTERT chromatin. Cancer Res 63(3): 689-695.
- Ducrest AL, Amacker M, Mathieu YD, Cuthbert AP, Trott DA, et al. (2001) Regulation of human telomerase activity: repression by normal chromosome 3 abolishes nuclear telomerase reverse transcriptase transcripts but does not affect c-Myc activity. Cancer Res 61(20): 7594-7602.
- Visakorpi T, Kallioniemi AH, Syvänen AC, Hyytinen ER, Karhu R, et al. (1995) Genetic changes in primary and recurrent prostate cancer by comparative genomic hybridization. Cancer Res 55(2): 342-347.
- Porkka Kati P, Visakorpi Tapio (2004) Molecular mechanisms of prostate cancer. European urology 45(6): 683-691.
- Karan Dev, Lin Ming-Fong, Johansson Sonny L, Batra Surinder K (2003) Current status of the molecular genetics of human prostatic adenocarcinomas. Int J Cancer 103(3): 285-293.
- Meeker AK (2006) Telomeres and telomerase in prostatic intraepithelial neoplasia and prostate cancer biology. Urol Oncol 24(2): 122-130.
- Botchkina GI, Kim RH, Botchkina IL, Kirshenbaum A, Frischer Z, et al. (2005) Noninvasive detection of prostate cancer by quantitative analysis of telomerase activity. Clin Cancer Res 11(9): 3243-3249.
- Graham MK, Meeker A (2017) Telomeres and telomerase in prostate cancer development and therapy. Nat Rev Urol 14(10): 607-619.
- Zheng SL, Stevens VL, Wiklund F, Isaacs SD, Sun J, et al. (2009) Two independent prostate cancer risk-associated Loci at 11q13. Cancer Epidemiol. Biomarkers Prev 18(6): 1815-1820.
- Chung CC, Ciampa J, Yeager M, Jacobs KB, Berndt SI, et al. (2011) Fine mapping of a region of chromosome 11q13 reveals multiple independent loci associated with risk of prostate cancer. Hum Mol Genet 20(14): 2869-2878.
- Chung CC, Boland J, Yeager M, Jacobs KB, Zhang X, et al. (2012) Comprehensive resequence analysis of a 123-kb region of chromosome 11q13 associated with prostate cancer. Prostate 72(5): 476-486.
- Nurminen Riikka, Wahlfors Tiina, Tammela Teuvo LJ, Schleutker Johanna (2011) Identification of an aggressive prostate cancer predisposing variant at 11q13. Int J Cancer International Journal of Cancer 129(3): 599-606.
- Nurminen Riikka, Rantapero Tommi, Wong Swee C, Fischer Daniel, Lehtonen Rainer, et al. (2016) Expressional profiling of prostate cancer risk SNPs at 11q13.5 identifies DGAT2 as a new target gene. Genes Chromosomes Cancer 55(8): 661-673.
- Linne H, Yasaei H, Marriott A, Harvey A, Mokbel K, et al. (2017) Functional role of SETD2, BAP1, PARP-3 and PBRM1 candidate genes on the regulation of hTERT gene expression. Oncotarget 8(37): 61890-61900.
- Schleutker Johanna, Baffoe-Bonnie Agnes B, Gillanders Elizabeth, Kainu Tommi, Jones Mary-Pat, et al. (2003) Genome-wide scan for linkage in finish Hereditary Prostate Cancer (HPC) families identifies novel susceptibility loci at 11q14 and 3p25-26. Prostate 57(4): 280-289.
- Barber L, Gerke T, Markt SC, Peisch SF, Wilson KM, et al. (2018) Family history of breast or prostate cancer and prostate cancer risk. Clin Cancer Res 24: 5910-5917.
- Lamy PJ, Tretarre B, Rebillard X, Sanchez M, Cenee S, et al. (2018) Family history of breast cancer increases the risk of prostate cancer: results from the EPICAP study. Oncotarget 9(34): 23661-23669.
- Beebe-Dimmer JL, Yee C, Cote ML, Petrucelli N, Palmer N, et al. (2015) Familial clustering of breast and prostate cancer and risk of postmenopausal breast cancer in the Women's Health Initiative Study. Cancer 121(8): 1265-1272.
- Motevalli A, Yasaei H, Virmouni SA, Slijepcevic P, Roberts T (2014) The effect of chemotherapeutic agents on telomere length maintenance in breast cancer cell lines. Breast Cancer Res Treat 145(3): 581-591.
© 2022. Terry Roberts. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






