- Submissions

Full Text
Novel Approaches in Cancer Study
Steric Influence on Anti-Cancer Activity of Phenyl Acridine Derivatives
Abhilash Thakur1, Mamta Thakur2*, Neelu Malviya3 and Brij Kishor Tiwari4
1Oxford International Institute, Shantanu Education Society, Indore, India
2Department of Applied Sciences, Symbiosis University of Applied Sciences, Indore, India
3Department of Chemistry, Maharani Laxmibai P.G. College, Indore, India
4Department of Chemistry, G.L. Bajaj Institute of Technology and Management, Greater Noida, India
*Corresponding author: Mamta Thakur, Department of Applied Sciences, Symbiosis University of Applied Sciences, Indore, India
Submission: June 01, 2020Published: August 18, 2020

ISSN:2637-773XVolume5 Issue1
Abstract
Present study represents the steric influence on the anti-cancer activity (DNA binding affinity) for the set of 27 Phenyl acridine derivatives. Quantitative structure activity methodology has been adopted to understand the steric influence on the DNA binding affinity of the compounds. Preliminary step of the method is the generation of structural, topological and physicochemical parameters. Relationship between these structural features and DNA binding affinity (logK) has been developed using multiple linear regression analysis.From the study it is observed that the parameters representing steric and volumetric parameters of the molecule are playing dominating role over the other parameters in characterization of the DNA binding of phenyl acridine derivatives as an anti-canceragent. Statistical analysis exhibits the dominance of approximate surface area and surface tension with hydrophobicity and polarizability factors in modeling of DNA binding affinity. QSAR models developed in present study are cross validated by variety of cross validation parameters. The model obtained in the study is useful to model a novel derivative in the series of Phenyl acridines.
Introduction
Acridine, (with the molecular formula C13H9N), a heterocyclic ring compound seen in crude anthracene. It was separated by treatment and shaking out with dilute sulphuric acid, and then precipitate out the sulphuric acid solution by the treatment of potassium dichromate, the resulting Acridine dichromate being rotten by ammonia [1]. Acridine and its homologues are very stable compounds of weakly basic character. These Acridine analogs can be crystallizes in needles which melt at 110° C and can be characterized or identify by its exasperating action on the skin, and by the blue fluorescence exhibits by the solutions of its salts [2,3]. Phenyl-acridine is the base of chrysaniline, which is the main constituent of the dyestuff phosphine (it is a by-product from the manufacturing of rosaniline) [4].
In addition to the chemical application acridines in the form of Phenyl Acridine exhibits wide spectrum of biological applications, important among these is its anti-cancer biological application. As it is well known fact that protein–DNA interactions shape the basis of different molecular biological processes required for the functioning of cells [5-7]. This form the basis of studies for the scientists to study the DNA-small molecule interactions, for example, interaction of DNA with different types of organic molecules [8-12], inorganic complexes [13-18], fluorescent materials [19,20] and different ions [21-23].
These studies are very helpful to understand the molecular basis of complex diseases and provide help to develop important drug candidates [24]. Most Acridine derivatives interact with DNA as inter-calator. Some bis-acridines bind to DNA through intercalation between consecutive nucleotides [25-27]. Binding of Acridine molecules interferes in the DNA function by blocking the DNA starter which is required by polymerases to synthesize RNA and DNA. In some of the cases the chemotherapeutic action of these Acridine is not restricted to DNA binding, even they also wield their effects on the inhibition of key enzymes such as topoisomerase, telomerase or polymerases. All these interactions are mostly non-covalent in the nature [27-29].
However, there are no reports on the biological activity of 9-phenyl acridine (PAc), although a number of synthetic derivatives of 9-arylacridine are known [30]. Molecular modelling studies show that these groups of drugs can act as inhibitors of topoisomerase and thus have potential to act as anticancer agents [31]. In this regard, the information about the mechanism of molecule DNA interaction is important to know. Some of the study shows their findings from computational modelling studies about the binding of PAc with DNA [32]. These studies are performed using biophysical techniques to establish the binding of the derivative with DNA and substantiated the findings with the help of molecular modelling studies. The Field of Quantitative Structure Activity Relationships (QSAR) is now well established [33] and this work has demonstrated that the biological activity of a set of 27 Phenyl Acridine derivatives acting via the same mode of action can be mathematically related to some simple physicochemical properties or molecular structure parameters [34,35]. These methods have been widely adopted in the pharmaceutical and agrochemical industries. In the present study efforts has been made to obtain an effective structure activity model which describes the structural requisitions within a Phenyl acridines to display its anti-cancer activity with improved efficacy. QSAR analysis to obtain mathematical model has been done using set of 27 Phenyl Acridine derivatives.
Methodology
In present study, methodology is based on aspect of Quantitative Structure Activity Relationship i.e, to develop mathematical model based on relation:
Φ = f (C)
where,
Φ = Biological property
C = Structural descriptor/ physicochemical properties
C used in present work is topological indices and physicochemical properties.
Biological activity
Biological activity analyzed in the present study is logK (Association constant) for the set of 27 Phenyl Acridine derivatives. The DNA binding activity is usually expressed in terms of association constant (logK) [36].
Topological/structural parameters
Second generation topological parameters Balaban branching index for hetro atoms using polarizability-based matrix and vander waal weighted matrix are used in present study [37].
Physicochemical properties tested in present investigation
The physicochemical properties describe various structural, physical & chemical assets of the compounds viz., size, polarizability, membrane transportation, inter and intra molecular forces vander Waal’s volume, weight etc. these physicochemical features plays the dominating role in deciding the biological activity or function of any molecule or chemical systems. Physicochemical parameter viz., Molar refractivity, Molar volume, Parachor, Index of refraction, Density, Polarizability and Surface Tension (ST) [38,39] is tested in present study, since Surface tension is found to play important role in binding affinity, therefore discussed here.
ST = (Pc / MV )4
Since ST is inversely proportional to MV, therefore, this is inverse steric effect.
Logarithm of octanol/water partition coefficient (logP)
The Octanol -Water partition coefficient is the ratio of concentration of specific compound in an Octanol/Water mixture.
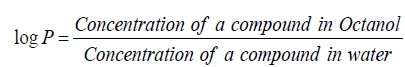
It shows hydrophobic interaction between a drug and a binding site at a receptor. It is used as a predictor of solute-membrane partitioning. All the above physicochemical properties are calculated using Chemsketch5.0 (www.acdlabs.com) while logP is calculated using available computer program.
Non-conventional physicochemical properties like approximate surface area (ASA), is the parameter used for the representation of size of the molecule with steric influence and tentative area covered with the bonding and overlapping of areas within the molecules [40].
Multiple Linear Regression (MLR)
MLR is an extension of simple linear regression by the inclusion of the extra independent variables
Y= ax1 + bx2 + ……. + constant.
The most familiar standard approaches to QSAR41 are based on multiple linear regression (MLR) and partial least squares (PLS) regression [41-43]. In present study linear mathematical models are developed to study Quantitative structure/Property Activity Relationship. Multiple linear regressions is used to develop models. Topological indices and physicochemical properties are used as independent variables to predict biological properties (dependent variable) as a DNA binding affinity log K of Pac derivatives. Bivariate and multivariate regression has been performed for finding out the best regression models. All those models having value of R below 0.50 are considered to be statistically insignificant.
Result and Discussion
As mentioned earlier in introduction that the study has been carried out on set of 27 Phenyl Acridine darivatives presented in Table 1 along with their DNA binding affinity. The parent or basic structure of the derivatives is presented in Figure 1. The derivatives shown in Table 1 are studied to explore the role of various structural and physicochemical properties, in DNA binding affinity of PAc. The descriptor which are involved in the QSAR model as a result of MLR analysis is depicted in Table 2 the QSAR model(s) obtained is shown as Eq 1-4 the best model can be utilize to design new derivative in a series, with more efficacy.
Figure 1: Basic structure of Phenyl Acridine derivatives.
Table 1: Substituents of Figure 1, Experimental log K, predicted log K of PAc derivatives and residual of Experimental and predicted lof K.
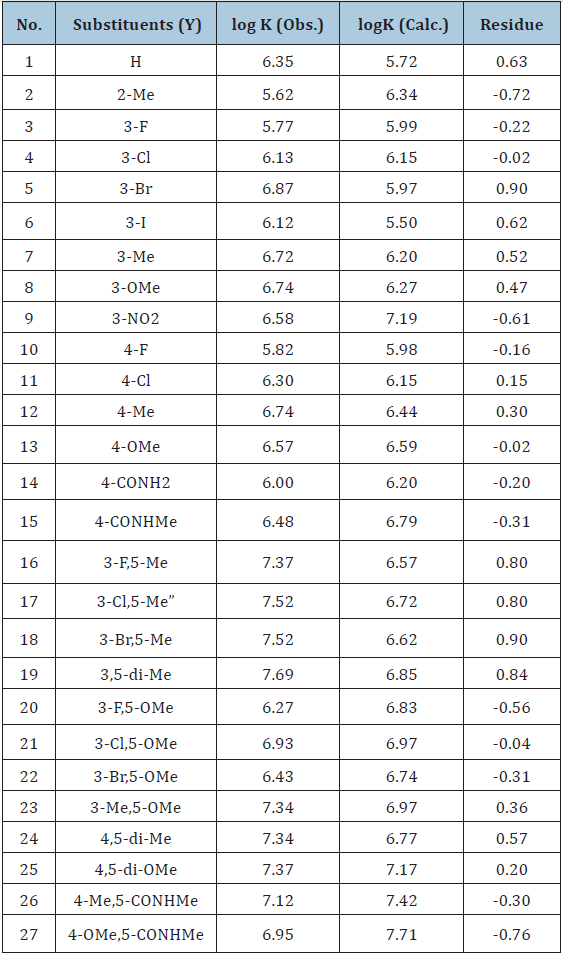
Table 2: Structural physicochemical, hydrophobic parameters tested in present study.
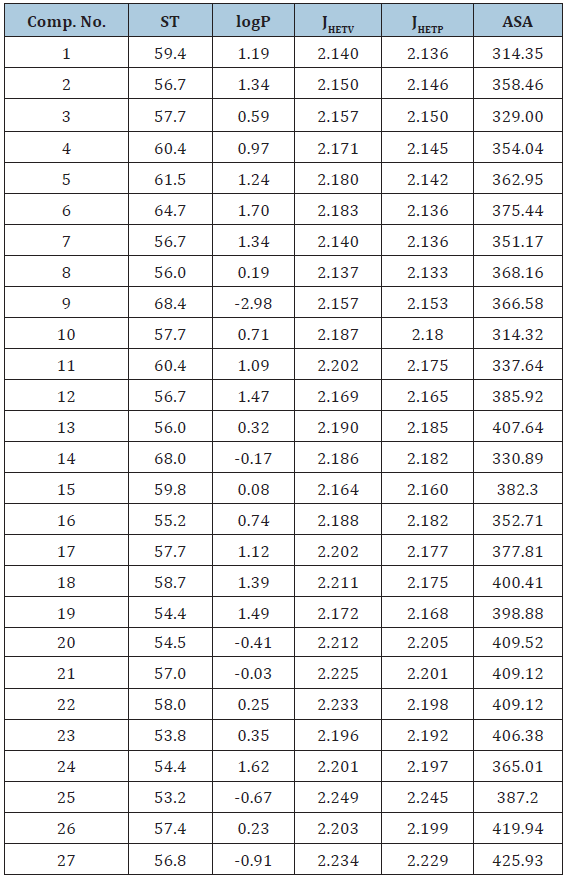
ST= Surface Tension; logP= Octanol /Water partition coefficient; JHETV= Balaban index for hetro atoms with vander wall weighted matrix; JHETP= Balaban index for hetro atoms with polarizability weighted matrix, ASA= Approximate Surface Area
However, Eq 5 is isoparametric to Eq 4 but with 26 derivatives, one derivative is considered as outlier out of 27 and hence excluded from the study being an exceptional derivative and Eq 5 has been developed with 26 PAc derivatives. In present study step up MLR method has been adopted in which mathematical model generation starts from univariate to bi, tri so on and so forth.
logK = 0.0089(±0.0032) ASA – 0.0418 (±0.0258) ST + 5.8021 Eq (1)
n = 27; Se = 0.4781; R = 0.6217; F = 7.562; Q = 1.300
logK = 0.0086(±0.0031) ASA – 0.0484 (±0.0257) ST + 3.2314(±2.268) JHETV+ 0.5786 Eq (2)
n = 27; Se = 0.4781; R = 0.6605; F = 5.934; Q = 1.381
logK = 0.0087(±0.0031) ASA – 0.0494 (±0.0257) ST + 3.3948(±1.9566) JHETP +0.5786 Eq (3)
n = 27; Se = 0.4592; R = 0.6764; F = 6.467; Q = 1.381
logK = 0.0056(±0.0035) ASA–0.0857(±0.0328) ST+7.5821(±3.1644) JHETP - 0.2628 (±0.1594) logP - 3.0732 Eq (4)
n = 27; Se = 0.4430; R = 0.7192; F = 5.892; Q = 1.623
logK = 0.0046(±0.0030) ASA–0.0944 (±0.0280) ST+8.2642(±2.6997) JHETP -0.2623 (±0.1355) logP - 3.2910 Eq (5)
n = 26; Se = 0.3766; R = 0.7846; F = 8.408; Q = 2.083
The magnitude of coefficients given in Eq(s) shows relative contribution of the corresponding parameter towards log K. in light of this fact the descending order of the influence of descriptors participating in final mathematical model Eq(5) is - JHEPT > log P > ST > ASA. JHEPT pertaining to the Balaban-type index from polarizability weighted distance matrix represent the polarizability distribution within the molecule, and positive coefficient of JHEPT in Eq 5 represent higher value of this parameter increases binding affinity of PAc derivative with DNA, which results in the formations of stable DNA-PAc derivative complex.
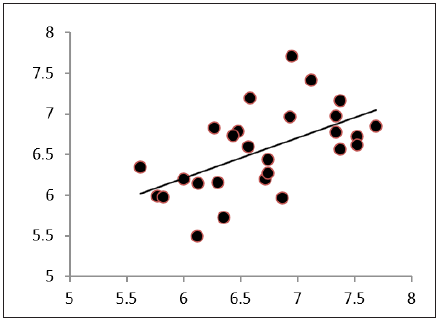
On contrary the negative coefficient of hydrophobic parameter log P represents the negative impact of hydrophobic contours on the binding affinity of the compounds towards DNA. Surface tension is also an important parameter in the Eq (5), which reflects tendency to shrink into the minimum surface area possible. Its negative coefficient indicates that PAc derivatives which undergoes least reduction in its volume during conformational changes, rotational, vibrational or translational motion are effective derivatives. Fourth parameter is approximate surface area of the compound which reflect the overall influential surface area of the molecule or an effective proximate area of the molecule, the positive coefficient of ASA indicate larger the influential area greater will be the DNA binding affinity of the molecule. Eq (5) is considered as a best mathematical model for the theoretical estimation of DNA binding affinity therefore log K of al 26 PAc derivatives has been estimated and presented with their residual values in Table 1,and represented graphically in Figure 2.
Figure 2: Graphical representation of correlation between observed and calculated logK.
Conclusion
The QSAR analysis performed on the set of 27 Phenyl Acridine derivatives subsequently leads to the conclusion that larger polarized area, lower hydrophobic contour, lesser surface tension and larger approximate surface area is favorable for the DNA binding affinity. The derivative best fitted in these four constrains will exhibits effective DNA binding and subsequently effective anticancer agent.
References
- Graebe C, Caro H (1871) Ueber acridin. Justus Liebigs Ann Chem 158: 265-281.
- Bernthsen A (1884) Die Acridine. Justus Liebigs Annalen der Chemie 224(1-2): 1-56.
- Gerard M, Kelly DP, Mack P (2001) Synthesis of acridine-based DNA bis-intercalating agents. Molecules 6(3): 230-243.
- Wilson WD, Shaikh Mizan, Farial T, Shijie Yao, Gerald Zon (1994) The interaction of intercalators and groove-binding agents with DNA triple-helical structures: The influence of ligand structure, DNA backbone modifications and sequence. Journal of Molecular Recognition 7(2): 89-98.
- Cate JH, Gooding AR, Podell E, Zhou K, Golden BL (1996) RNA tertiary structure mediation by adenosine platforms. Science 273 (5282): 1696-1699.
- Leitinger N, Wesierska-Gadek J (1993) ADP-ribosylation of nucleolar proteins in HeLa tumor cells. Journal of Cellular Biochemistry 52(2): 153-158.
- Duncan HB (1988) Characterization of the fluorescence of the antitumor agent, mitoxantrone. Biochimica Et Biophysica Acta (BBA) - Gene Structure and Expression 949(1): 132-137.
- Austin CA, Marsh KL (1998) Eukaryotic DNA topoisomerase Iiβ. Bioessays 20(3): 215-226.
- Pommier Y (1993) DNA topoisomerase I and II in cancer chemotherapy: Update and perspectives. Cancer Chemother Pharmacol 32(2): 103-108.
- Padgett D, Brandner W, Stapelfeldt K, Strom S, Terebey S, et al. (1999) Hubble space telescope/nicmos imaging of disks and envelopes around very young stars. The Astronomical Journal 117: 1490-1504.
- Willmore E, Frank AJ, Padget K, Tilby MJ, Austin CA (1998) Etoposide targets topoisomerase IIα and IIβ in leukemic cells: Isoform-specific cleavable complexes visualized and quantified in situ by a novel immunofluorescence technique. Mol Pharmacol 53: 78-85.
- Errington J, Mandelstam J (1986) Use of a lacZ gene fusion to determine the dependence pattern of sporulation operon spoIIA in spo mutants of Bacillus subtilis. J Gen Microbiol 132(11): 2967-2976.
- Errington F, Willmore E, Tilby MJ, Li L, Li G, et al. (1999) Murine transgenic cells lacking DNA topoisomerase IIβ are resistant to acridines and mitoxantrone: Analysis of cytotoxicity and cleavable complex formation. Molecular Pharmacology 56(6): 1309-1316.
- Jehn U, Heinemann V (1993) Phase- II study of treatment of refractory acute leukemia with intermediate-dose cytosine arabinoside and amsacrine. Annals of Hematology 66(3): 131-134.
- Turnbull DH, Ramsay JA, Shivji GS, Bloomfield TS, From L, et al. (1996) Ultrasound backscatter microscope analysis of mouse melanoma progression. Ultrasound Med Biol 22(7): 845-853.
- Moreland RB, Langevin GL, Singer RH, Garcea RL, Hereford LM (1987) Amino acid sequences that determine the nuclear localization of yeast histone 2B. Mol Cell Biol 7(11): 4048-4057.
- Finley JP, Ögelman H, Kiziloglu U (1992) Rosat observations of PSR 0656+14: A pulsating and cooling neutron star. Astrophysical Journal Letters 394: 21.
- Anne Huff, Robert Ward, Kenneth NK (1990) Mutational alteration of the breakage/resealing subunit of bacteriophage T4 DNA topoisomerase confers resistance to antitumor agent m-AMSA. Molecular & Genral Genetics 221(1): 27-32.
- Wasserman RA, Wang JC (1994) Mechanistic studies of amsacrine-resistant derivatives of DNA topoisomerase II. Implications in resistance to multiple antitumor drugs targeting the enzyme. The Journal of Biological Chemistry 269(33): 20943-2095.
- Todd AC, Lisa MW, Neil PS, Anne MV, Miles AF, et al. (2005) Inhibition of drug-resistant mutants of ABL, KIT, and EGF receptor kinases. Proc Natl Acad sci USA 102(31): 11011-1106.
- Ming-Sheng, Wang C, Miloslav B (1992) Two independent amsacrine-resistant human myeloid leukemia cell lines share an identical point mutation in the 170kDa form of human topoisomerase II. Journal of Molecular Biology 223(4): 837-843.
- Hinds M, Karl D, Janice M, Elizabeth A, Ruud J, et al. (1991) Identification of a point mutation in the topoisomerase II gene from a human leukemia cell line containing an amsacrine-resistant form of topoisomerase II. Cancer Res 51(17): 4729-4731.
- Fisher LM, Cullen ME, Wyke AW, Kuroda R (1989) Cloning and characterization of a DNA gyrase A gene from Escherichia coli that confers clinical resistance to 4-quinolones. Antimicrob Agents Chemother 33(6): 886-894.
- Chrysoula L, Gilroy L, Gary PW, Pauline H, Ian G, et al. (2007) Differential selection of acridine resistance mutations in human DNA topoisomerase IIβ is dependent on the acridine structure. Mol Pharmacol 71(4): 1006-1014.
- Christian B (2000) Topoisomerase I poison and suppressors as anticancer drugs. Current Medicinal Chemistry 7(1): 39-58.
- Rita Ghosh, Sudipta B, Angshuman B, Dipankar Das, Somnath G (2010) Chemotherapeutic potential of 9-phenyl acridine: Biophysical studies on its binding to DNA. European Biophysics Journal 39(8): 1243-1249.
- Imad IH, Graham GS, Roger DW (2001) Ligand binding to oligonucleotides. Methods in Molecular Biology 163: 379-391.
- Stupina VA, Wang JC (2005) Viability of Escherichia coli topA mutants lacking DNA topoisomerase. J Biol Chem 280: 355-360.
- Bugreev DV, Nevinsky GA (2009) Structure and mechanism of action of type IA DNA topoisomerases. Biochemistry (Moscow) 74(13): 1467-1481.
- Bhowmic S, Bagchi A, Ghosh R (2008) Molecular modeling studies of some 9 arylacridine to elucidate their possible role in topoisomerase I inhibition. International Journal of Integrative Biology 2(1): 8-14.
- Gentry AC, Juul S, Veigaard C, Knudsen BR, Osheroff N (2011) The geometry of DNA supercoils modulates the DNA cleavage activity of human topoisomerase I. Nucleic Acids Res 39(3): 1014-1022.
- Abhilash Thakur, Mamta Thakur, Nitika K, Ashok J, Ashok G (2004) Application of topological and physicochemical descriptors: QSAR study of phenylamino-acridine derivatives. Arkivoc 10(1): 36-43.
- Leo A, Hansc, C, Church C (1969) Comparison of parameters currently used in the study of structure-activity relationships. J Med Chem 12(5): 766-771.
- Martin YC (1978) Quantitative drug design, medicinal research series. Marcel Dekker, New York, USA, Volume 8.
- Hansch C, Leo A (1979) Substituent constants for correlation analysis in chemistry and biology. J Wiley and Sons, Brisbane, Australia.
- Corwin H, Alka K, Rajni G, Hua G (2001) Chem-bioinformatics and QSAR: A review of QSAR lacking positive hydrophobic Terms. Chem Rev 101(3): 619-672.
- Balaban AT (1986) Chemical graphs. part 48. topological index j for heteroatom-containing molecules taking into account periodicities of element properties. MATCH (Commun Math Chem) 21: 115-122.
- Abhilash Thakur (2005) QSAR Study on benzenesulfonamide DNA binding affinity: Physicochemical approach using surface tension. ARKIVOC 14: 49-53.
- M Thakur, Abhilash T, PV Khadikar, Supuran CT (2005) QSAR Study On pKa vis-à-vis physiological activity of sulfonamides: A dominating role of surface tension (inverse steric parameter). Bioorg Med Chem Lett 15(1): 203-209.
- Mamta T, Abhilash T, Lokendra O (2014) Surface area grid in modeling of Anti-HIV activity of TIBO Derivatives. Int J of Res & Dev in Pharmacy and Life Sciences 3(3): 983-992.
- Trinajstic N (1992) In chemical graph theory; CRC Press: Boca Raton, FL, USA, pp. 225-273.
- Basak SC, Grunwald GD, Niemi GJ (1977) Use of graph-theoretic geometrical molecular descriptors in structure-activity relationships. In from chemical topology to three-dimensional geometry. Plenum Press, New York, USA, pp.73-116.
- Lalit K, Mamta T, Abhilash T, Amit KT, Nagendrappa G (2017) Modeling of optimum dose for carboquinone derivatives as anti-leukemic agent. Indian Research Communication 11(1): 1-11.
© 2020. Mamta Thakur. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






