- Submissions

Full Text
Novel Approaches in Cancer Study
Identifying Biomarkers for Diffuse Large B-Cell Lymphoma Subtypes
Prashanthi Dharanipragada and Nita Parekh*
Centre for Computational Natural Sciences and Bioinformatics, India
*Corresponding author:Nita Parekh, Centre for Computational Natural Sciences and Bioinformatics, Hyderabad, India
Submission: August 23, 2019;Published: September 11, 2019

ISSN:2637-773XVolume3 Issue2
Abstract
Diffuse Large B-cell Lymphoma (DLBCL) is a highly heterogeneous cancer of B cells. Apart from cell-of-origin, genetic variations have been observed to contribute towards heterogeneity leading to different pathogenic mechanisms and overall survival outcomes. Various classification schemes have been proposed that may aid in risk stratification and developing new therapeutics for those who fail frontline therapy. This mini review highlights the significance of genetic variations as biomarkers for DLBCL and ease in extending it to clinical setting.
Keywords: Genetic variations; Biomarkers; DLBCL; Classification; Pathways; Therapeutics
Background
Diffuse Large B-cell Lymphoma (DLBCL) is the most common lymphoid malignancy in adults, accounting for ~35% of Non-Hodgkin’s lymphoma cases worldwide. Several classification systems have been proposed based on shared morphology, immunophenotype, genetic alterations, clinical outcomes, etc., to decipher the mechanisms of pathogenesis and design suitable therapy. Classification of DLBCL into cell-of-origin (COO) subtypes, Germinal Centre B-cell (GCB) and Activated B-cell (ABC), has been traditionally defined as low- and highrisk patient groups respectively when treated with chemotherapy [1]. Several previous studies have shown a diverse set of genetic and epigenetic factors affecting few key pathways in each subtype. For example, the GCB subtype is mostly characterized with alterations in chromatinmodifying enzymes, activated PI3K pathway, disruption of Gα migration pathway components and frequent structural variants of BCL2 gene, while ABC subtype is often associated with increased NF-κB activity, altered BCR signalling, perturbed terminal B cell differentiation, etc. The differences in the genetic profiles of the two subtypes have been associated with differential response to treatment, with ABC subtype exhibiting poorer outcomes compared to GCB subtype when treated with standard rituximab, cyclophosphamide, doxorubicin (Adriamycin), vincristine (Oncovin) and prednisone (R-CHOP) immuno-chemotherapy. Moreover relapsed/refractory DLBCL is observed in ~30-40% of the treated cases.
Traditional methods for subtype identification
From previous clinical studies, it is evident that correct diagnosis of the subtype (GCB/ ABC) at an early stage is important for effective personalized treatments. Gene expression profiling (GEP) is one of the earliest techniques to classify the DLBCL subtypes and several studies have indicated a varying set of differentially expressed genes having predictive and prognostic value. For example, overexpression of genes BCL6 and NFKB1 are exclusively associated with GCB and ABC subtypes, respectively [2]. As GEP is not routinely available in clinical use, immunohistochemical (IHC) algorithms have been developed to predict the patient’s cancer subtype and outcome. Among these, Visco-Young algorithm is a recent method that considers the expression of CD10, FOXP1 and BCL6 and has shown higher concordance with GEP results for DLBCL classification with prognostic significance [3]. Though frequently used in clinical practice, these traditional approaches suffer from certain limitations. The mRNA quality of tumour cells considered for GEP is generally deteriorated due to formalinfixed, paraffin-embedded (FFPE) tissues, resulting in a large number of false predictions. The IHC methods may show reaction bias (e.g., specimen fixation) and interpretation bias (e.g., selection of antibody panels, types and clones and assessment of results) leading to a number of false positives and false negatives [4]. Also, the exact cause of differential expression of genes and hence, subtype-specific mechanisms of tumorigenesis cannot be comprehended through these conventional approaches.
Genetic variations as potential biomarkers
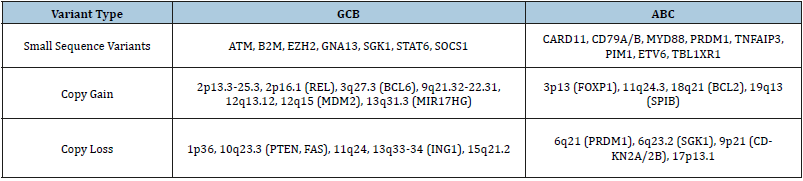
Genetic Variations, ranging from single nucleotide variations to large structural alterations (insertions and deletions, inversions, translocations and copy number variations (CNVs)) have been shown to play a potential role in the predisposition of several cancers, including DLBCL. Recently, genome-wide studies of DLBCL using next-generation sequencing (NGS) have revealed a large number of somatic mutations that may contribute to the heterogeneous behaviour of DLBCL [5]. Few NGS studies have also analysed copy number alterations to understand the molecular heterogeneity in DLBCL subtypes [6]. Table 1 summarizes few known genetic aberrations associated with GCB and ABC subtypes that may serve as diagnostic and/or prognostic markers. In a recent study [7], integrated analysis of targeted next-generation sequencing of 106 genes and genomic copy number alterations (CNAs) in 150 DLBCL patients revealed differential profile of mutations and CNVs, resulting in altered pathogenic pathways in the two subtypes. Importantly, the prognostic value of NOTCH signalling pathway (NOTCH1/NOTCH2 mutations) and deletion of TP53/CDKN2A involved in cell cycle were shown to be associated with poor outcome across both the subtypes. Genetic variations and their associated immuno-oncogenic pathways identified are often targetable, and a number of drugs are already undergoing clinical trials. For example, somatic mutations in histone acetyltransferases, CREBBP and EP300, lead to DLBCL lymphomagenesis (mostly found in GCB subtype). In such cases, inhibition of histone deacetylase (HDAC) indicates a promising therapeutic strategy due to their precise selectivity and tolerable levels of toxicity in humans [8].
Table 1:Genes carrying genetic variations (SNVs and CNVs) that have been characterized as molecular biomarkers for the GCB and ABC DLBCL subtypes listed.
Recently, novel classifications of DLBCL subtypes have been proposed based on genetic variants. For instance, genomic and transcriptomic analysis of 574 DLBCL biopsy samples revealed four distinct subtypes, viz. MCD (co-occurrence of MYD88L265P and CD79B mutations), BN2 (BCL6 fusions and NOTCH2 mutations), N1 (NOTCH1 mutations), and EZB (EZH2 mutations and BCL2 translocations) [9]. In another study [10] on a cohort of 304 primary DLBCL cases identified five distinct molecular subsets, including a novel GCB/ABC independent group (C0), based on genetic variations. Some of the key genetic variations in clusters include C1: BCL6 structural variations, C2: TP53 mutations, CDKN2A and RB1 deletions, C3: BCL2 mutation and PTEN deletion and C4: alterations in histone genes. These subtypes exhibited distinct molecular profiles, activation/inhibition of different pathogenic pathways and varied response to immuno-chemotherapy, affecting the overall outcome of patients. Clearly, a high genetic complexity is observed and traditional COO classification categorizing patients into GCB or ABC subtypes is not sufficient for patient stratification and recognizing potential targets for new intervention strategies. Developing gene panels that can be easily used in a clinical setting may provide a solution, e.g., ‘Lymphopanel’ that identifies mutations in 34 genes across 8 immuno-oncogenic pathways [11].
Conclusion
Genetic variations, which includes sequence (SNVs and short INDELs) and structural variations (CNVs, translocations and inversions), are frequently observed in DLBCL, and act as potential diagnostic and prognostic biomarkers. Functional analysis of these variants would enable in understanding the molecular basis of heterogeneity, patient risk stratification and designing treatment regimens. Although several studies have identified a diverse set of genetic variations associated with the pathogenesis of DLBCL subtypes, a complete spectrum of genetic lesions in these subtypes and their associated pathways still remain to be elucidated. The major challenge is in finding the clinical relevance of recurrent genomic alterations. There clearly is a need for comprehensive resource annotating associations of tumour alterations with pathways, potential drug targets, and their relevant effect on anticancer drugs. Low sequencing costs and fast generation of sequence data has now made it possible to take genetic variant detection, annotation and association from the phenotype to clinical setting.
References
- Scott DW (2015) Cell-of-origin in diffuse large B-cell lymphoma: Are the assays ready for the clinic? Am Soc Clin Oncol Educ Book: e458-e466.
- Puvvada S, Kendrick S, Rimsza L (2013) Molecular classification, pathway addiction, and therapeutic targeting in diffuse large B-cell lymphoma. Cancer Genet 206(7-8): 257-265.
- Visco C, Li Y, Xu-Monette ZY, Miranda RN, Green TM, et al. (2012) Comprehensive gene expression profiling and immunohistochemical studies support application of immunophenotypic algorithm for molecular subtype classification in diffuse large B-cell lymphoma: A report from the International DLBCL Rituximab-CHOP Consortium Program Study. Leukemia 26(9): 2103-2113.
- Matos LL, Trufelli DC, de Matos MGL, da Silva Pinhal MA (2010) Immunohistochemistry as an important tool in biomarkers detection and clinical practice. Biomark Insights 5: 9-20.
- Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, et al. (2012) Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci USA 109(10): 3879-3884.
- Sebastián E, Alcoceba M, Martín-García D, Blanco Ó, Sanchez-Barba M, et al. (2016) High-resolution copy number analysis of paired normal-tumor samples from diffuse large B cell lymphoma. Ann Hematol 95(2): 253-262.
- Karube K, Enjuanes A, Dlouhy I, Jares P, Martin-Garcia D, et al. (2018) Integrating genomic alterations in diffuse large B-cell lymphoma identifies new relevant pathways and potential therapeutic targets. Leukemia 32(3): 675-684.
- Havas AP, Rodrigues KB, Bhakta A, Demirjian JA, Hahn S, et al. (2016) Belinostat and vincristine demonstrate mutually synergistic cytotoxicity associated with mitotic arrest and inhibition of polyploidy in a preclinical model of aggressive diffuse large B cell lymphoma. Cancer Biol Ther 17(12): 1240-1252.
- Schmitz R, Wright GW, Huang DW, Johnson CA, Phelan JD, et al. (2018) Genetics and pathogenesis of diffuse large B-cell lymphoma. New England Journal of Medicine 378(15): 1396-1407.
- Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, et al. (2018) Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med 24(5): 679-690.
- Dubois S, Viailly PJ, Mareschal S, Bohers E, Bertrand P, et al. (2016) Next-generation sequencing in diffuse large B-cell lymphoma highlights molecular divergence and therapeutic opportunities: A LYSA Study. Clin Cancer Res 22(12): 2919-2928.
© 2019 Nita Parekh. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






