- Submissions

Full Text
Modern Research in Dentistry
Clinical Implications of Single Nucleotide Polymorphisms in Oral Cancer
Hetal Damani Shah1 and Dhananjaya Saranath2*
1Department of Biological Sciences, Sunandan Divatia School of Science, SVKM’s NMIMS (Deemed-to-be) University, India
2Cancer Patients Aid Association, Dr. Vithaldas Parmar Research & Medical Centre, India
*Corresponding author: Dhananjaya Saranath, Cancer Patients Aid Association, Dr. Vithaldas Parmar Research & Medical Centre, Sumer Kendra, Worli, Mumbai, India
Submission: April 26, 2022;Published: June 03, 2022

ISSN:2637-7764Volume7 Issue2
Abstract
Background: Oral cancer is a high incidence cancer with well-established risk factors including tobacco,
alcohol, areca nut, and HR-HPV 16/18. Additional risk factors such as Single Nucleotide Polymorphisms
(SNPs) associated with genetic susceptibility, disease progression and prognosis. A comprehensive
review of clinically relevant SNPs is synopsized in the current article.
Methods: The PubMed repository was primarily used to retrieve articles using a combined keyword
search. We selected 81 eligible studies examining association of SNPs with oral cancer, published in peer
reviewed journals, between January 2000 and March 2022, with a focus on SNPs and prediction of oral
cancer survival indicating response to therapy.
Results: Twelve SNPs including ERCC5 rs17655, ERCC1 rs735482, TP53 rs1042522, MDM2 rs2279744,
MTHFR rs1801133, MTHFR rs1801131, MSH2 rs3732183, MLH1 rs1800734, FADS1 rs174549, XPD
rs13181, XPD rs1799793, and TGFBR1 rs33438 emerged as potential SNP markers for prediction of oral
cancer survival and response to therapy.
Conclusion:
The risk-loci may be indicated potential biomarkers for early oral cancer detection, better patient prognosis and also serve as molecular targets for novel therapies.Keywords: SNPs; Oral Cancer; Prognosis; Survival; Recurrence; Response to therapy
Introduction
Cancer incidence and mortality rates are increasing globally with an estimated 19.3 million new cancer cases and 9.96 million cancer deaths as per Globocan 2020 [1]. Globally, oral cancer accounts 377,713 new case annually and 177,757 deaths; and India marks the highest incidence of oral cancer worldwide with 135,929 new cases contributing 35.98% to the global oral cancer burden [1]. Oral cancer is a multifactorial disease with well-established risk factors including tobacco, areca nut, alcohol and high-risk HPV 16/18 [2]. Further, the molecular landscape of oral cancer encompasses somatic mutations, deregulated expression, epigenetic regulation via DNA methylation, histone modification and microRNAs, and genomic variants [3]. Despite technological advances and improvements in diagnosis and treatment modalities, the 5-year survival rate of oral cancer patients is poor [4]. Thus, there is need for relevant additional oral cancer specific biomarkers for early diagnosis, prognosis and identification of therapeutic targets for better clinical outcome. In the last two decades, the genomic constitution of an individual has been associated with cancer susceptibility and implicated in disease progression, patient survival, response to therapy and prognosis, as well as identification of therapeutic targets [5-8].
Single nucleotide polymorphisms (SNPs), contribute almost 90% of genomic variants in individuals, and have emerged as critical modulators of host disease susceptibility, drug responses and prognosis [9]. Studies from our group defined mutations and polymorphisms of biologically functional genes including oncogenes, tumor suppressor genes, apoptotic genes, receptor genes and signal transduction genes in oral cancer [10- 15]. With advances in technology, high throughput microarray based whole genome expression analysis and whole genome association studies (GWAS), confirmed by nucleotide sequencing and allele specific oligonucleotide assays for examination of the genome, we identified several SNPs associated with increased or decreased risk of developing oral cancer [16-22]. Several global studies have highlighted the role of specific SNPs in oral cancer predisposition and oral cancer [17,23-28]. Further SNP data was compiled using PubMed repository and web of science to provide better understanding of the complex interplay of SNPs with oral cancer progression and prognosis.
Methodology
The following criteria is generally used in association studies. A systematic literature search of the PubMed repository for enlisting genetic association studies of SNPs with oral cancer, conducted globally and published in the English language between January 1990 and March 2022, using keywords in titles and abstracts e.g., ‘snps’ OR ‘SNP’ OR ‘Polymorphism’ and ‘Oral Cancer’ guiding the search. Additionally, the reference list of all short-listed studies is manually meeting our inclusion criteria and manually searched using the google search engine and web of science. All the relevant full-length articles are checked, and important data elements were noted with relevant details.
Result
Prototype result interpretation is given in this section. SNP studies including 459 articles were retrieved and yielded 94 publications for complete review of full-text articles. If necessary additional research articles are identified from a manual search of the reference list. Articles may be excluded if not relevant to the search. A total of 81 eligible original global research studies were included in the final review (Figure 1). The reviewed studies enabled delineation of specific SNPs e.g., in oral cancer 12 SNPs in ten critical genes were associated with patient survival and response to therapy as summarized in Table 1 and Figure 2. Several independent studies have reported the prognostic significance of ERCC5 rs17655 and ERCC1 rs735482, MLH1 rs1800734, TP53 rs1042522, MDM2 rs2279744, MTHFR rs1801133 and MTHFR rs1801131 with poor therapeutic response in oral cancer patients (Table 1) [29-33]. Whereas MSH2 rs3732183, MLH1 rs1800734, FADS1 rs174549, XPD rs13181, XPD rs1799793, TGFBR1 rs33438 are associated with better response (Table 1) [34-36].
Figure 1: Flow chart describing the search and selection of clinically relevant literature on SNPs associated with oral cancer.
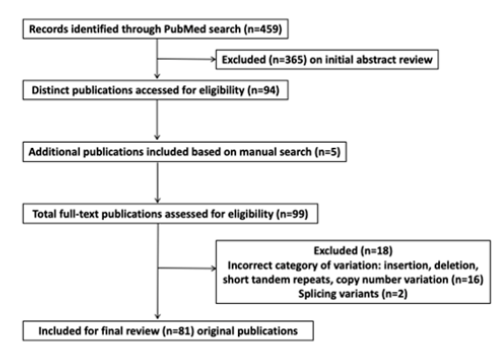
Table 1: SNPs associated with Increased Risk, Survival and Response to Therapy in Oral Cancer.

Discussion
Oral carcinogenesis is a complex multi-step process with defined hallmarks including selective growth and proliferative advantage, altered stress response favoring overall survival, vascularization, invasion and metastasis, metabolic rewiring, an abetting microenvironment, and immune modulation [37,38]. The twelve identified SNPs in ten genes associated with critical cellular functions involved in DNA synthesis, damage, repair, methylation, oncogene activation, proliferation and apoptosis are highlighted in Figure 2. These include ERCC5 rs17655, ERCC1 rs735482, TP53 rs1042522, MDM2 rs2279744, MTHFR rs1801133, MTHFR rs1801131, MSH2 rs3732183, MLH1 rs1800734, FADS1 rs174549, XPD rs13181, XPD rs1799793, and TGFBR1 rs33438 associated with survival outcomes in response to therapy in oral cancer patients [29-36].
Figure 2: Schematic representation of specific genes highlighting critical SNPs in the functional pathways of DNA Synthesis, Repair, Damage, Methylation, Apoptosis and Proliferation. The SNPs are indicated with response to therapy in oral cancer.
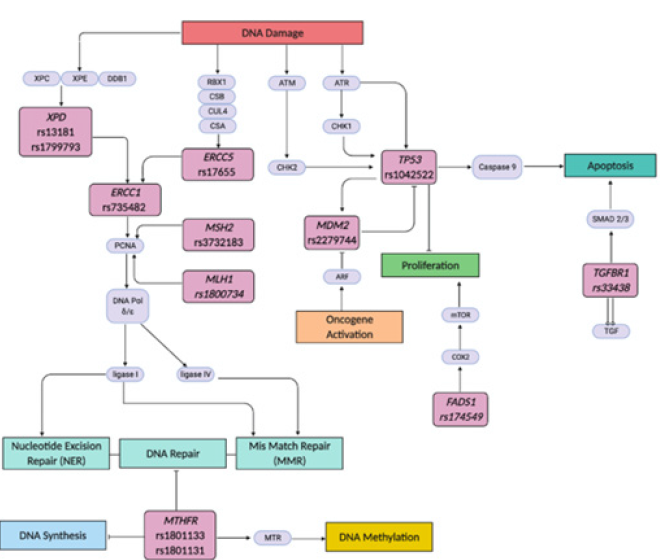
Chemo-resistance is implicated in the reduction of therapeutic efficacy, consequently affecting treatment outcome. Besides, Polymorphisms in DNA repair genes may alter protein function, affecting the anticancer effects of therapeutic agents and patients’ response to chemo/radiotherapy. Senghore et al. [33] identified key SNPs in nucleotide excision repair (NER) pathway associated with clinical outcome in oral squamous cell carcinoma (OSCC) patients treated with concurrent chemoradiotherapy (CCRT) [33]. The NER pathway genes play a major role in the repair of DNA damaged by chemotherapeutic agents and radiotherapy. The excision repair cross-complementation genes including ERCC1, ERCC2 or XPD, and ERCC5. Further, xeroderma pigmentosium complementation group A (XPA) and C (XPC) encode proteins involved in the NER pathway [33]. The carriers of the XPC rs2228000 TT genotype showed a borderline increased risk of poor OS (HR-1.81). Further, the CC genotypes of ERCC5 rs17655 and ERCC1 rs735482 (HR=1.54 and 1.65, respectively) were associated with an increased risk or worse DFS and increased recurrence (HR=2.60, 95% CI = 1.11-6.09) (Table 1) [33].
ERCC1 is responsible for DNA incision whereas ERCC5 coded DNA endonuclease causes excision and repair of UV-induced DNA damage thus the CC genotypes of ERCC5 rs17655 and ERCC1 rs735482 may increase the DNA-repair capacity of cancer cells,leading to increased susceptibility to recurrence implying that CCRT may not be beneficial, and alternative treatments should be considered in the patients with these SNPs [33]. On the contrary, a study by Mahimkar et al. [35] involving the NER pathway XPD genes demonstrate XPD rs1799793 and XPD rs13181 variant alleles, independently as well as in combination, as important predictors of clinical outcome in radiotherapy treated OSCC patients with XPD rs13181 associated with increased RFS while XPD rs1799793 corelated with increased RFS as well as DSS [35]. Since XPD protein is critically involved in the transcription factor IIH (TFIIH) complex in NER process, the XPD SNPs may confer survival advantage in Oral cancer patients by reducing DNA damage proficiency and thereby corelate with better treatment outcomes [35].
Senghore et al. [36] have demonstrated association of DNA mismatch repair (MMR) pathway genes MLH1 rs1800734 and MSH2 rs3732183 polymorphisms with treatment outcome in oral cancer patients [36]. The wild type genotypes ‘GG’ of mismatch repair pathway genes MSH2 rs3732183 and MLH1 rs1800734 exhibit higher DFS and are associated with better survival in Oral cancer patients treated with adjuvant CCRT (Table 1). Thus, the wild type of genotype is associated with improved prognosis in oral cancer patients [36]. MSH2 rs3732183 and MLH1 rs1800734 may alter the capacity of individuals to repair DNA damage induced by radiotherapeutic and chemotherapeutic agents. Since the MSH2 and MLH1 play significant roles in the repair process by recognizing the DNA damage caused by, the polymorphic allelic variants may alter the function [36]. As reported by Lin et al. [29], the polymorphic MLH1 rs18007 is associated with poor patient prognosis. The impact of MLH1 polymorphism on treatment outcome in oral cancer patients with primary surgery with or without adjuvant radiotherapy, indicated the association of MLH1 rs1800734 ‘AA’ (the homozygous SNP genotype) with poor prognosis with lower OS and DFS [29]. Thus, MLH1 rs1800734 may be predictive of poor prognosis in advanced stages Oral cancer patients with postoperative adjuvant radiotherapy indicating alternate treatment approach (Table 1) [29]. An interesting study by Kuroda and colleagues examined the relationship between SNPs in tumor suppressor TP53 and prognosis in oral cancer.
The TP53 polymorphism in codon72 on the 4th exon Arg (CGC) to Pro (CCC), results in poor prognosis in patients with TP53 rs1042522 in non-smoking individuals with the CC genotype with shorter post-treatment survival compared to the GC genotype (p=0.06) (Table 1) [39]. The wild type of GG genotype codes for Arginine producing wild type TP53 reported to induce apoptosis with faster kinetics and suppress transformation more efficiently than the Proline coding CC genotype [40]. Further, combination of MDM2 rs2279744 G/G and TP53 rs1042522 G/G polymorphism was associated with poorer prognosis (OS, HR-2.42, DFS, HR-2.90) and influence the outcome of advanced OSCC treated with adjuvant radiation predicting poor treatment response (Table 1) [32].
Folate deficiency influences the risk of cancer either by inducing mis incorporation of uracil into DNA leading to chromosomal breaks and mutations and/or by causing aberrant DNA methylation, resulting in altered expression of critical protooncogenes and tumor suppressor genes. Besides, polymorphisms in the Methylenetetrahydrofolate reductase (MTHFR) gene, coding the key reductase enzyme in folate metabolism, are implicated in oral cancer. The most common allele variants of MTHFR include C677T (MTHFR rs1801133) and A1298C (MTHFR rs1801131), that lead to amino acid substitutions (Ala222Val and Glu429Ala) resulting in thermolabile enzyme with reduced activity [31]. Sailasree [31] evaluated the influence of MTHFR rs1801133 and MTHFR rs1801131 on oral cancer patient survival. MTHFR rs1801133 polymorphic ‘TT’ genotype showed improved survival as compared to patients with wild type ‘CC’ genotype (RR=0.56, P=0.378). However, MTHFR rs1801131 ‘CC’ and ‘AC+CC’ showed an increased risk for treatment failure and poor survival when compared with the wild ‘AA’ genotype (HR=4.27, P=0.001) (Table 1) [31] attributed to impaired enzyme activity causing folate deficiency and consequent DNA methylation and genome instability.
SNPs associated with favorable patient outcomes and better response to therapy include the fatty acid desaturase 1 (FADS1) and the transforming growth factor beta receptor 1 (TGFBR1) genes [34]. FADS1 is the key rate-limiting enzyme of polyunsaturated fatty acids (PUFAs), which convert dihomo-gamma-linolenic acid (DGLA) to arachidonic acid (AA). AA and prostaglandin E2 (PGE2) modulate tumor microenvironment, promote carcinogenesis by increased proliferation and angiogenesis via the activation of PI3K-AKT signaling and mTOR signaling. The knockdown of FADS1 inhibited cancer growth, migration, and enhanced the cytotoxicity of chemotherapeutic agents [41]. FADS1 rs174549 is determined to be a potentially independent and favorable factor in predicting oral cancer PFS HR-0.52 (95% CI: 0.29-0.93) for patients with CCRT (Table 1) [34]. TGFBR1 plays a critical role in the TGF‐β/ SMAD signaling pathway vital in the development and progression of cancer by regulating cellular proliferation and differentiation [42]. The TGFBR1 rs33438 polymorphism is associated with a low risk of death due to oral cancer in codominant (AG vs AA: HR=0.55, 95% CI=0.35-0.88) and dominant (GG+AG vs AA: HR=0.57, 95% CI=0.38-0.87) models. Moreover, better disease specific survival (DSS) (GG+AG vs AA: HR=0.50, 95% CI=0.29-0.85) was reported in patients given radiotherapy. There also existed a positive multiplicative interaction on DSS between the polymorphism of TGFBR1 rs334348 and radiotherapy (P=0.001) (Table 1) [42]. TGFBR1 rs334348 polymorphism may influence the progression of oral cancer by regulating the levels of TGFBR1 and inhibiting its function of phosphorylating Smad‐2 and Smad‐3 and consequent increased cell proliferation, a hallmark of cancer [42].
Conclusion
Despite extensive research and technological advances in the last two decades, oral cancer mortality and morbidity rates have not improved. Thus, it is imperative to assess the vast array of factors that contribute to the prognosis of patients with oral cancer. Since the outcomes vary from one individual to another, the genomic variants represented as single nucleotide polymorphisms with prognostic relevance may serve as critical assessment factors of patient risk and aid the selection of optimal treatment strategies. With emerging targeted therapies, the SNP-genes also serve as therapeutic targets for small drug like molecules. Specific SNPs may act as predictive and prognostic markers enabling the clinicians to mitigate the risk associated with treatment response to conventional radiation and chemotherapy. The current review highlights twelve SNPs including ERCC5 rs17655, ERCC1 rs735482, TP53 rs1042522, MDM2 rs2279744, MTHFR rs1801133, MTHFR rs1801131, MSH2 rs3732183, MLH1 rs1800734, FADS1 rs174549, XPD rs13181, XPD rs1799793, and TGFBR1 rs33438 in oral cancer prognosis acting as potential SNP markers for prediction of oral cancer survival and response to therapy. However, it is important to note that individual SNPs reviewed are present in low penetrance genes, thus larger independent prospective studies with clinically relevant subgroups and endpoints are warranted.
References
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, et al. (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71(3): 209-249.
- Bagan J (2009) Oral squamous cell carcinoma overview. Oral Oncol 45(4-5): 301-308.
- Souza WD, Saranath D (2017) OMICS, oral cancer molecular landscapes, and clinical practice. Omi A J Integr Biol 21(12): 689-703.
- Schmitz S, Ang KK, Vermorken J, Haddad R, Suarez C, et al. (2014) Targeted therapies for squamous cell carcinoma of the head and neck: Current knowledge and future directions. Cancer Treat Rev 40(3): 390-404.
- Shah HD, Saranath D, Murthy V (2020) A molecular dynamics and docking study to screen anti-cancer compounds targeting mutated p53. J Biomol Struct Dyn 40(6): 2407-2416.
- Damani Shah H, Saranath D, Das S, Kharkar P, Karande A (2019) In-silico identification of small molecules targeting H-Ras and in-vitro cytotoxicity with caspase-mediated apoptosis in carcinoma cells. J Cell Biochem 120(4): 5519-5530.
- Li CC, Shen Z, Bavarian R, Yang F, Bhattacharya A (2018) Oral Cancer: Genetics and the Role of Precision Medicine. Dent Clin North Am 62(1): 29-46.
- Jackson M, Marks L, May GHW, Wilson JB (2018) The genetic basis of disease. Essays Biochem 62(5): 643-723.
- Andrew AS, Gui J, Sanderson AC, Mason RA, Morlock E V, et al. (2009) Bladder cancer SNP panel predicts susceptibility and survival. Hum Genet 125(5-6): 527-539.
- Saranath D, Panchal R, Nair R, Mehta A, Sanghavi V, et al. (1990) Restriction fragment length polymorphism of the L-myc gene in oral cancer patients. Br J Cancer 61(4): 530-533.
- Bhoite LT, Saranath D, Nair R, Deo MG, Sanghavi V, et al. (1993) H-ras-1 restriction fragment length polymorphism in normal individuals and oral cancer patients in India. J Oral Pathol Med 22(7): 298-302.
- Saranath D, Panchal RG, Deo MG, Sanghvi V, Mehta AR (1994) Restriction fragment length polymorphisms of the human N-myc gene in normal healthy individuals and oral cancer patients in India. Indian J Biochem Biophys 31(3): 177-83.
- Tandle A, Sanghavi V, Saranath D (2000) Infrequent loss of heterozygosity at adenomatous polyposis coli gene locus in Indian oral cancers. Cancer Lett 157(2): 155-60.
- Tandle AT, Sanghvi V, Saranath D (2001) Determination of p53 genotypes in oral cancer patients from India. Br J Cancer 84(6): 739-742.
- Bhatnagar R, Dabholkar J, Saranath D (2012) Genome-wide disease association study in chewing tobacco associated oral cancers. Oral Oncol 48(9): 831-835.
- D’Souza W, Pradhan S, Saranath D (2017) Multiple single nucleotide polymorphism analysis and association of specific genotypes in FHIT, SAMD4A, and ANKRD17 in Indian patients with oral cancer. Head Neck 39(8): 1586-1595.
- Multani S, Saranath D (2016) Genotypic distribution of single nucleotide polymorphisms in oral cancer: global scene. Tumor Biol 37(11): 14501-14512.
- Multani S, Saranath D (2016) Single nucleotide polymorphisms and risk of oral cancer: Indian case-control study. J Clin Cell Immunol 7(4): 1-5.
- Multani S, Pradhan S, Saranath D (2016) Gene polymorphisms and oral cancer risk in tobacco habitué Tumor Biol 37(5): 6169-6176.
- Damani Shah H, Saranath D, Pradhan S (2020) Single nucleotide polymorphisms in transcription factor genes associated with susceptibility to oral cancer. J Cell Biochem 121(2): 1050-1060.
- Multani S, Shah H, Saranath D (2019) Transforming growth factor beta receptor 2 single-nucleotide polymorphism association with oral cancer and in silico identification of small drug-like molecules as inhibitors to transforming growth factor Beta-2 receptor. Biomed Res J 6(1): 25.
- Yete S, Pradhan S, Saranath D (2017) Single nucleotide polymorphisms in an Indian cohort and association of CNTN4, MMP2 and SNTB1 variants with oral cancer. Cancer Genet 214-215: 16-25.
- Zhang ZJ, Hao K, Shi R, Zhao G, Jiang GX, et al. (2011) Glutathione S-Transferase M1 (GSTM1) and Glutathione S-Transferase T1 (GSTT1) null polymorphisms, smoking, and their interaction in oral cancer: a HuGE review and meta-analysis. Am J Epidemiol 173(8): 847-857.
- Liao G, Wang Y, Zhou YQ, Li TW, Zeng DQ, et al. (2014) Host genetic susceptibility to oral cancer: evidence from meta-analyses and pooled analyses. Oral Dis 20(7): 644-649.
- Pereira A, Dias do Carmo E, Dias da Silva M, Blumer Rosa L (2012) Matrix metalloproteinase gene polymorphisms and oral cancer. J Clin Exp Dent 4(5): e297-301.
- Marichalar Mendia X, Rodriguez Tojo MJ, Acha Sagredo A, Rey Barja N, Aguirre Urizar JM (2010) Oral cancer and polymorphism of ethanol metabolising genes. Oral Oncol 46(1): 9-13.
- Zygogianni AG, Kyrgias G, Karakitsos P, Psyrri A, Kouvaris J, et al. (2011) Oral squamous cell cancer: early detection and the role of alcohol and smoking. Head Neck Oncol 3: 2.
- Hernando Rodriguez M, Rey Barja N, Marichalar Mendia X, Rodriguez Tojo MJ, Acha Sagredo A, et al. (2012) Role of cytochrome P-450 genetic polymorphisms in oral carcinogenesis. J Oral Pathol Med 41(1): 1-8.
- Lin LH, Lin MW, Mar K, Lin CS, Ji DD, et al. (2014) The hMLH1-93G>A promoter polymorphism is associates with outcomes in oral squamous cell carcinoma patients. Ann Surg Oncol 21(13): 4270-4277.
- Huang SF, Chen IH, Liao CT, Wang HM, Liou SH, et al. (2009) Combined effects of MDM2 SNP 309 and p53 mutation on oral squamous cell carcinomas associated with areca quid chewing. Oral Oncol 45(1): 16-22.
- Sailasree R, Nalinakumari KR, Sebastian P, Kannan S (2011) Influence of methylenetetrahydrofolate reductase polymorphisms in oral cancer patients. J Oral Pathol Med 40(1): 61-66.
- Tu HF, Chen HW, Kao SY, Lin SC, Liu CJ, et al. (2008) MDM2 SNP 309 and codon 72 polymorphisms are associated with the outcome of oral carcinoma patients receiving postoperative irradiation. Radiother Oncol 87(2): 243-252.
- Senghore T, Chien HT, Wang WC, Chen YX, Young CK, et al. (2019) Polymorphisms in ERCC5 rs17655 and ERCC1 rs735482 genes associated with the survival of male patients with postoperative oral squamous cell carcinoma treated with adjuvant concurrent chemoradiotherapy. J Clin Med 8(1): 33.
- Chen F, He B, Yan L, Qiu Y, Lin L, et al. (2017) FADS1 rs174549 polymorphism may predict a favorable response to chemoradiotherapy in oral cancer patients. J Oral Maxillofac Surg 75(1): 214-220.
- Mahimkar MB, Samant TA, Kannan S, Tulsulkar J, Pai PS, et al. (2012) Polymorphisms in GSTM1 and XPD genes predict clinical outcome in advanced oral cancer patients treated with postoperative radiotherapy. Mol Carcinog 51 Suppl 1: E94-E103.
- Senghore T, Wang WC, Chien HT, Chen YX, Young CK, et al. (2019) Polymorphisms of mismatch repair pathway genes predict clinical outcomes in oral squamous cell carcinoma patients receiving adjuvant concurrent chemoradiotherapy. Cancers (Basel) 11(5): 598.
- Fouad YA, Aanei C (2017) Revisiting the hallmarks of cancer. Am J Cancer Res 7(5): 1016-1036.
- Tanaka T, Ishigamori R (2011) Understanding carcinogenesis for fighting oral cancer. J Oncol 2011: 603740.
- Kuroda Y, Nakao H, Ikemura K, Katoh T (2007) Association between the TP53 codon72 polymorphism and oral cancer risk and prognosis. Oral Oncol 43(10): 1043-1048.
- Thomas M, Kalita A, Labrecque S, Pim D, Banks L, et al. (1999) Two polymorphic variants of wild-type p53 differ biochemically and biologically. Mol Cell Biol 19(2): 1092-1100.
- Yang X, Xu Y, Brooks A, Guo B, Miskimins KW, et al. (2016) Knockdown delta-5-desaturase promotes the formation of a novel free radical byproduct from COX-catalyzed ω-6 peroxidation to induce apoptosis and sensitize pancreatic cancer cells to chemotherapy drugs. Free Radic Biol Med 97: 342-350.
- Chen L, Chen F, Wang X, Chen Q, Lin J, et al. (2020) Prognostic value of transforming growth factor beta receptor 1 polymorphisms in patients with oral cancer. J Oral Pathol Med 49(2): 137-144.
© 2022 Dhananjaya Saranath. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






