- Submissions

Full Text
Modern Research in Dentistry
Detection of Staphylococcus aureus in the Pulp of an Endocarditis Patient
Oumarou Hama H1,2, Gouriet F1,5, Catherine JH3,4,5, Habib G1,5, Raskin A3,4,5, Hadj Said M3,5, Lan R3,4,5, Aboudharam G2,3,5, Drancourt M1,2,5 and Terrer E2,3,5*
1IHU Méditerranée Infection, Marseille, France
2Aix-Marseille Université, IRD, MEPHI, IHU Méditerranée Infection, Marseille, France
3Aix-Marseille Université, UFR Odontologie, Marseille, France
4Aix-Marseille Université, Anthropologie bioculturelle Droit Ethique Santé, Marseille, France
5APHM, Hôpital Timone, Marseille, France
*Corresponding author: Elodie Terrer, Aix-Marseille University, MEPHI, IHU Méditerranée-Infection 19-21 Boulevard Jean Moulin, 13005 Marseille, France
Submission: July 08, 2020;Published: July 20, 2020

ISSN:2637-7764Volume5 Issue2
Abstract
Objective: Dental pulp is acknowledged to be an organic tissue sample on which the microbiological diagnosis of blood-borne pathogens, including those responsible for infectious endocarditis, can be based.
Method: Molecular detection of aureus was performed in the dental pulp extracted from one tooth collected in a patient firmly diagnosed with S. aureus infectious endocarditis.
Result: We report on one patient diagnosed with Staphylococcus aureus endocarditis in whom aureus DNA was further detected by PCR in the dental pulp. We advocate not throwing away extracted teeth, appropriate microbial investigations of which may reveal bacteraemic pathogens not otherwise detectable.
Keywords: Bacteremia; Tooth; MALDI TOF MS; Microbiology; Diagnosis; Cardiology
Introduction
Dental pulp is acknowledged to be a suitable organic tissue upon which to base the microbiological diagnosis of blood-borne pathogens, including those responsible for infectious endocarditis [1,2]. Non-exposed dental pulp can be invaded by bacteria with predominance of aero-intolerant bacteria of the genera Eubacterium, Propionibacterium and Actinomyces [3]. Furthermore, molecular approaches have made it possible to detect the DNA of Coxiella burnetii, a pathogen responsible for endocarditis [4] in the dental pulp of experimentally-infected guinea pigs [5], as well as HIV DNA in a seropositive patient [6]. In addition, one case of Bartonella quintana bacteraemia has been detected by PCR in the dental pulp sample collected from a patient who had been diagnosed with B. quintana bacteraemia six months previously but who was not longer bacteraemic when the tooth was extracted [1]. Infectious endocarditis (IE) is mainly caused by streptococci, staphylococci and enterococci and, rarely, by fungi [7,8]. Staphylococcus aureus (S. aureus) is one of the pathogens most frequently involved in blood-borne infections, including IE [9,10]. In this paper, we report on one patient diagnosed with S. aureus endocarditis in whom S. aureus DNA was further detected by PCR in the dental pulp.
Materials
In April 2019, a 54-year-old patient was admitted to the emergency department of the Timone hospital (Marseille, France) with fever, confusion and tetraparesia. A cerebral scan found multiple left hypodense fronto-parietal and left occipital lesions. Blood and the cerebrospinal fluid cultured methicillin-susceptible S. aureus. Empiric antibiotic therapy combining amoxicillin, gentamycin and acyclovir was then changed for cefazolin, 12g/day and clindamycin 600mg four times/day. A transthoracic echocardiography found vegetation on the native bicuspid aortic valve and a moderate aortic insufficiency. The patient was then diagnosed with S. aureus infectious endocarditis. A total body scan found bilateral renal, splenic and hepatic embolisms. Brain magnetic resonance imaging found several right frontal, left occipital, temporal, thalamic and bilateral cerebellar hyper signals with haemorrhagic organisation in the right frontal and occipital lesion. Five days later, the patient presented with septic shock and was admitted to the cardiologic intensive care unit. Antibiotic therapy was changed for intravenous sulfamethoxazole 4,800mg/day, clindamycin 1,800mg/day, gentamycin 160mg/day and rifampicin 1,800mg/day.
Laboratory tests showed leucocytosis at 19G/L, haemoglobin at 10.4g/dL, platelets at 180G/L, and protein C-reactive at 95.5mg/L. 18fluorodeoxyglucose-positron emission tomography/computed tomography showed multiple hypermetabolic foci in the lymph nodes, liver, muscles and bones. Spinal magnetic resonance imaging showed a lumbar L4-L5 and cervical C5-C6 spondylodiscitis. Initial clinical and biological evolution was favourable, however, three weeks later the cardiac lesion worsened with severe aortic valve insufficiency. Cardiac surgery was performed and the patient underwent an aortic bio prosthesis. Culture of the explanted cardiac valve remained sterile and PCR-based tests for the pan bacterial 16S RNA gene, Enterococcus faecalis, Enterococcus faecium, S. aureus, Coxiella burnetii and Bartonella spp. were negative. Histological examination of the explanted valve showed a valvular tissue largely destroyed by a dense, non-specific and polymorphic inflammatory reaction, with many neutrophils.
As part of the routine management of patients with IE, 13 teeth with infectious foci of endodontic or periodontal origin were extracted using amoxicillin antibiotic prophylaxis according to the recommendations of the Haute Autorité de Santé (HAS) and the Agence Nationale de Sécurité du Médicament (ANSM). After obtaining the patient’s informed consent, the teeth were microbiologically investigated in line with advice from the IHU Mediterranean Infection Ethics Committee (Advice, 05/29/2018). The evolution of the patient was favourable at the six-month follow-up consultation. Six of the 13 extracted teeth (n°13, n°24, n°25, n°35, n°36, n°37) were used for bacteriology investigation. Teeth were decontaminated with 1% chlorhexidine (MP Biomedicals, Illkirch, France), washed with UltraPure™ DNase/RNase-Free Distilled Water (Invitrogen, Illkirch, France), and fractured longitudinally (Table 1). The pulp was extirpated from each tooth using a sterile excavator and cut into three fragments collected in sterile tubes, as previously described [11]. For the bacterial culture, one pulp fragment was crushed in a tube containing 30µL of sterile phosphate-buffered saline (PBS), inoculated on two plates of COS agar (bioMérieux, Mercy l’Etoile, France), and incubated at 37 °C without and/or with 5% CO2 for 48 hours. Colonies were identified using matrix-associated laser desorption ionization/time of flight mass spectrometry (MALDI TOF MS), as previously described [12]. Two pulp fragments from teeth n°13, n°24 and n°37 were used for S. aureus quantitative qPCR and fast culturomics, as previously described [13].
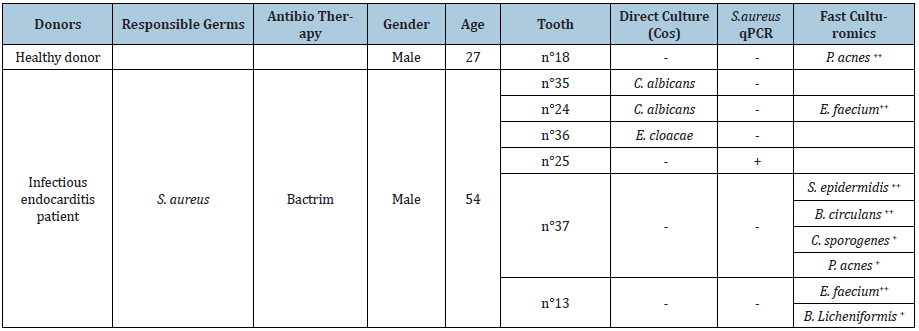
Table 1: Results and identification of bacteria found by direct culture, real-time PCR (qPCR) and fast culturomics of one negative control patient and one patient with infectious endocarditis. (“+” denotes a positive result; “-” denotes a negative result).
Total DNA was extracted as follows: the pulp fragment was mixed with glass powder and 200μL of G2 buffer and shaken using the FAST prep at 400rpm for one minute. 20μL of proteinase K 25mg/mL and 20μL of phage T4 were then added and incubated at 56 °C overnight with shaking. 200μL of the mixture was then eluted in a volume of 50μL, following the standard procedure of the EZ1 Advanced XL Extraction Kit (QIAGEN, Hilden, Germany). Negative controls included sterile PBS and the dental pulp collected from a wisdom tooth (n°18) from a healthy donor. Extracted DNA was incorporated into a real-time PCR targeting a fragment of 116bp of the S. aureus nucA 2 gene (F: 5 'GTTGTGGATGGTGATACATTTATTGC 3', P: 5 '6FAM- AGGCTTATAGGGGTTGATACGCCAGAAACGG 3', R: 5' CCAAATGGTTGTACAGGCGTATTC 3’) [unpublished data]. The PCR mixture incorporated 15µL of mix and 5µL of extracted DNA. The amplification reaction was performed in a CFX96TM real-time PCR system (Bio-Rad, Marnes-la-Coquette, France) using the following protocol: one cycle of 50 °C for two minutes for the UDG action then five minutes initial denaturation at 95 °C followed by 40 cycles of denaturation at 95 °C for five seconds and a hybridisation-elongation step of 30 seconds at 60 °C.
The fast culturomics protocol was applied, as described previously [13] for teeth n°13, n°24 and n°37 and tooth n°18 from a negative control healthy donor. In short, pulps were cultured in an anaerobic atmosphere under three conditions including two pre-incubation steps. A direct inoculation of the pulps on COS agar and Yeast Casitone Fatty Acids (YCFA) agar was carried out followed by two preincubation inoculations, one in liquid YCFA medium and the other one in a bottle of anaerobic blood culture enriched with 5% rumen fluid and 5% sheep blood. Pre-incubation samples were sub-cultured on the COS and YCFA agar media at three hours, six hours, nine hours, 24 hours, three days, seven days and 10 days using cascade dilution series. Bacteria were identified by MALDI-TOF-MS after 48 hours of incubation.
Methods and Result
Teeth n°13, n°25 and n° 37 were negative for the standard bacterial culture, teeth n°24 and n°35 cultured Candida albicans, and tooth n°36 cultured Enterobacter cloacae, despite decontamination. Fast culturomics yielded Enterococcus faecium from teeth n°24 and n°13 under at least two conditions and Bacillus licheniformis was also identified in tooth n°13. Further, Staphylococcus epidermidis, Bacillus circulans, Clostridium sporogenes and Propionibacterium acnes were identified in tooth n°37 while Propionibacterium acnes was the only germ identified in the healthy donor. S. aureus DNA was amplified in tooth n°25 by targeting the nucA 2 gene and no amplification was observed with the extraction blank, reaction mix and healthy donor. Furthermore, the control of DNA extraction and the presence of genetic material from the six samples was validated by phage T4 PCR and albumin PCR.
Discussion
Molecular detection of S. aureus was performed on the dental pulp extracted from one tooth collected from a patient definitively diagnosed with S. aureus infectious endocarditis. The results are presented in Table 1. The absence of contamination of the extracted dental pulp was ensured by the meticulous cleaning of the tooth with UltraPure™ DNase/RNase-Free Distilled Water after using chlorhexidine before the extirpation of the dental pulp. Molecular detection was confirmed by the absence of amplification of the extraction blank and the reaction mix. To our knowledge, this is the second case in which the DNA of the pathogen responsible for infectious endocarditis was found in the dental pulp of the patient. The first case was the detection of Bartonella quintana in a patient who had been bacteraemic six months previously [1].
Aerobic culture of two of the six teeth yielded C. albicans and E. cloacae whereas culture of the negative control wisdom tooth remained negative, and three teeth from the patient also remained negative. Cultured pathogens may differ from tooth to tooth in the same individual. Moreover, yeasts have already been reported in the dental pulp despite previous decontamination of the manipulated tooth and the presence of antifungal in the culture medium [14]. E. faecium isolated in two of three teeth under at least two rapid culturomics conditions is one of the most prevalent pathogens responsible for IE [7,15]. Its detection in the patient reported on here may result from past E. faecium bacteraemia.
A few weeks prior to tooth extraction, the patient had S. aureus bacteraemia which had remained undocumented in the extracted cardiac valve by culture and PCR-based methods after three weeks of appropriate antibiotic therapy. In this context, S. aureus was detected by PCR in the dental pulp, illustrating the interest of microbial investigations of the dental pulp. This is, therefore, the second ever report of detection of a bacteraemic pathogen in the dental pulp, following our previous report of a similar observation in a patient with B. quintana endocarditis and bacteraemia [1]. These clinical observations may be more than simply fortuitous, as an experimental guinea pig model of C. burnetii bacteraemia also showed the pathogen to be detectable in the dental pulp [5]. These combined observations suggest that dental pulp may be a sanctuary for bacteraemic bacteria in which their elimination is delayed.
Conclusion
In conclusion, we advocate using extracted teeth to search for bacteraemic pathogens rather than disposing of them as is currently the case, as microbiological investigations of the dental pulp may offer diagnoses that are not otherwise achievable; including the very particular situation of post-mortem search for the cause of death in which PCR-based and culture-based investigations may reveal deadly septicaemia and endocarditis.
References
- Aboudharam G, Fournier PE, Drancourt M, Raoult D, Foucault C, et al. (2004) Molecular detection of Bartonella quintana DNA in the dental pulp of a homeless patient. Eur J Clin Microbiol Infect Dis. 23(12): 920-922.
- La VD, Aboudharam G, Raoult D, Drancourt M (2008) Dental pulp as a tool for the retrospective diagnosis of infectious diseases. In: Raoult D, Drancourt M (Eds.), Paleomicrobiology Past Human Infections, Springer, Berlin (Heidelberg), Germany, pp. 175-196.
- Hoshino E, Ando N, Sato M, Kota K (1992) Bacterial invasion of non-exposed dental pulp. Int Endod J 25(1): 2-5.
- Jang YR, Song JS, Jin CE, Ryu BH, Park SY, et al. (2018) Molecular detection of Coxiella burnetii in heart valve tissue from patients with culture-negative infective endocarditis. Medicine (Baltimore) 97(34): e11881.
- Aboudharam G, Lascola B, Raoult D, Drancourt M (2000) Detection of Coxiella burnetii DNA in dental pulp during experimental bacteremia. Microb Pathog 28(4): 249-254.
- Glick M, Trope M, Pliskin ME (1989) Detection of HIV in the dental pulp of a patient with AIDS. J Am Dent Assoc 119(5): 649-650.
- Blanchard V, Pagis B, Richaud R, Moronval F, Lutinier R, et al. (2020) Infective endocarditis in French Polynesia: Epidemiology, treatments and outcomes. Arch Cardiovasc Dis 113(4): 252-262.
- Ellis M (1997) Fungal endocarditis. J Infect 35(2): 99-103.
- Laupland KB (2013) Incidence of bloodstream infection: a review of population-based studies. Clin Microbiol Infect 19(6): 492-500.
- Fowler VG Jr, Miro JM, Hoen B, Cabell CH, Abrutyn E, et al. (2005) Staphylococcus aureus Endocarditis: A Consequence of Medical Progress. JAMA 293(24): 3012-3021.
- Drancourt M, Aboudharam G, Signoli M, Dutour O, Raoult D (1998) Detection of 400-year-old Yersinia pestis DNA in human dental pulp: An approach to the diagnosis of ancient septicemia. Proc Natl Acad Sci U S A 95(21): 12637-12640.
- Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, et al. (2009) Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis 49(4): 543-551.
- Naud S, Khelaifia S, Mbogning D, Dione N, Lagier JC, et al. (2020) Proof of concept of culturomics use of time of care. Cellular and Infection Microbiology.
- Perry BC, Zhou D, Wu X, Yang FC, Byers MA, et al. (2008) Collection, cryopreservation, and characterization of human dental pulp-derived mesenchymal stem cells for banking and clinical use. Tissue Eng Part C Methods 14(2): 149-156.
- Fernández Guerrero ML, Goyenechea A, Verdejo C, Roblas RF, de Górgolas M (2007) Enterococcal endocarditis on native and prosthetic valves: A review of clinical and prognostic factors with emphasis on hospital-acquired infections as a major determinant of outcome. Medicine (Baltimore) 86(6): 363-377.
© 2020 Terrer E. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






