- Submissions

Full Text
Modern Concepts & Developments in Agronomy
First Report of Tomato Spotted Wilt Orthotospovirus Infecting Polygonum Multiflorum in China
Qin Zhuo 1,2#, Xue Zheng2#, Shaozhi Zhang2, Xiaoman Ai3, Xiaofang Zhang3, Lihua Zhao2* and Zhongkai Zhang2**
1College of Agriculture, Yunnan University, Kunming 650504, Yunnan, China
2Biotechnology and Germplasm Resources Institute, Yunnan Academy of Agricultural Sciences; Yunnan Provincial Key Lab of Agricultural Biotechnology; Kunming 650205, Yunnan, China
3Southwest Forestry University, Kunming 650224, Yunnan, China
*Corresponding author:Lihua Zhao, Biotechnology and Germplasm Resources Institute, Yunnan Academy of Agricultural
Sciences; Yunnan Provincial Key Lab of Agricultural Biotechnology; Kunming 650205,Yunnan, China
Zhongkai Zhang, Biotechnology and Germplasm Resources Institute, Yunnan Academy of Agricultural Sciences; Yunnan Provincial Key Lab of Agricultural Biotechnology;
Kunming 650205, Yunnan, China
Submission: April 12, 2023;Published: May 05, 2023

ISSN 2637-7659Volume13 Issue 1
Abstract
Tomato Spotted Wilt orthotospovirus (TSWV) is a member of the Orthotospovirus genus. It has extensive host and causes serious disease in many crops. In May 2021, virus-like symptoms such as chlorosis, mottling, leaf yellowing and necrosis were observed on Polygonum multiflorum (hereinafter as P. multiflorum) leaves in Kunming, Yunnan Province, China. Spherical viral particles with a diameter of 80-120nm were observed by TEM.RT-PCR and DNA sequencing analysis were performed, and the results showed that the sequence of N gene for this virus shared 99.20% identity with TSWV isolate from Shandong isolate (acc. no. MN861982.1).The obtained 777-bp consensus sequence named TSWN-N was deposited in GenBank (acc. no. OP800910).Phylogenetic tree analyses suggested that TSWV obtained from P. multiflorum plant samples clustered together into a large clade from Korea. Koch’s postulate was performed, and the virus was re-amplied from the inoculated plants. This is the first report of TSWV on P. multiflorum in China and provides early warning of virus disease on medicinal materials.
Keywords:Tomato spotted wilt orthotospovirus; Polygonum multiflorum; Electron microscopy; RT-PCR
Introduction
Polygonum multiflorum (hereinafter as P. multiflorum) is a perennial herb that belongs to the genus Pleuropterus genus and Polygonaceae family [1]. It is a famous traditional Chinese medicine and as a tuber has been used for treating disease such as blackening hair, anti- aging, anti-hyperlipidemia, antioxidant, anti-inflammatory, anticancer, hepatoprotection, cardio-protection and improving age-related cognitive dysfunction [2]. Owing to its high medicinal value, P. multiflorum usually is distributed mainly in Sichuan, Yunnan, Guizhou and southern Shanxi and Gansu in China [1]. Diseases often occur in the planting process and cause serious economic losses. The most widespread disease on P. multiflorum is ceitocybe bescens, rust disease, leaf spot disease and some fungal diseases or insect pests [3-5]; However, there are few reports about the virus that infects P. multiflorum. Polygonum ringspot virus (PolRSV), a recently described tospovirus [6], has been reported only on the weed species Polygonum dumetorum and Polygonum convolvolus in different geographic areas in Italy. So far, there have been no reports about the tomato spotted wilt virus infecting P. multiflorum [7,8]. In this paper, symptomless P. multiflorum plants leaves were received from professionally cultivated fields for regular control of virus infection in May 2021. TEM and the molecular investigations indicated that this virus was TSWV. This work was done to fulfill Koch’s postulate and verify TSWV is able to infect P. multiflorum plants after artificial inoculation in fields.
Materials and Methods
Samples
Surveys were conducted in May 2021 in Kunming, Yunnan Province, China. Symptoms were observed on individual P. multiflorum of the same variety displaying virus-like symptoms including foliar viral symptoms consisting of chlorosis, mottling, leaf yellowing and necrosis. The five sample leaves of P. multiflorum were collected from diseased parts of plants and stored in a refrigerator at -80 °C on the same day for RT-PCR.
Transmission electron microscopy
The virion morphology and size of the virus in P. multiflorum were confirmed by Transmission Electron Microscopy (TEM). Following the methods of introduction, negative staining was performed for infected and healthy leaves and was observed under a transmission electron microscope with an accelerating voltage of 80kV [9].
RT-PCR amplification
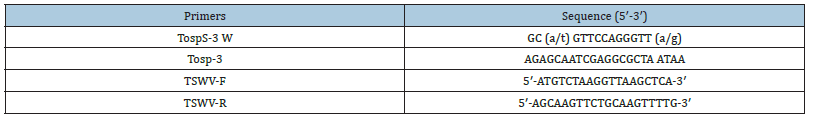
Total RNA was extracted from symptomatic and symptomless leaves using Trizol reagent (Nanjing, Norvezan Biotechnology) according to the manufacturer’s protocol. The total RNA extracted above was reverse transcribed to complete the first-strand cDNA synthesis according to the HiScriptIII 1st Strand cDNA Synthesis Kit (Nanjing, Norvezan Biotechnology). The primers were designed by primer 5 and Table 1. PCR reaction system of 25μL: 10×PCR reaction buffer (including Mg2+) 2.5μL, dNTP Mix 2μL, 10μmol/L upstream and downstream primers each of 0.5μL, rTaq DNA polymerase 0.25μL, cDNA 1.25μL, ddH2O 18μL. Amplification conditions: 94 °C pre-denaturation 30s, 98 °C denaturation 10s, 53 °C annealing 30s (TospS-3 W/Tosp-3 primer annealing temperature 53 °C, TSWV-F / TSWV-R primer annealing temperature 58 °C), 72 °C extension for 45s, 35 cycles, 72 °C further extension for 10 min.
Target gene clones and sequencing analysis
The target fragment DNA was recovered and purified. The recovered products were connected to D18-T vector using the fast DNA ligation kit. The ligation products were transferred into E.coli DH5αcompetent cells. The positive clones were selected and sent to Beijing Qingke Biotechnology Co., Ltd. for sequencing, and the sequencing results were analyzed by BLAST alignment analysis.
Phylogenetic analyses
Phylogenetic analyses were conducted using MEGA version 6.0; The maximum likelihood that the (ML) tree was constructed with 1000 replicates as the guide value to evaluate the reliability of the resulting tree [10].
Result
TEM detection
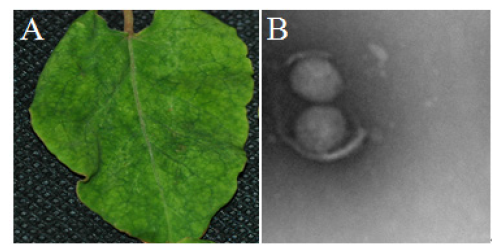
In May 2021, virus-like symptoms on leaves consisting of chlorosis, mottling, leaf yellowing and necrotic were observed and appeared on approximately 80% of P. multiflorum in the fields in Kunming, Yunnan (Figure 1A). The five samples with symptoms and symptomless were taken for Transmission Electron Microscopy (TEM) detection using negative staining [9], respectively. The results revealed spherical particles of 80-120 nm in diameter, similar to Orthotospovirus (Figure 1B).
Figure 1:Electron micrograph of virus particles found in P. multiflorum leaves. A. Pictures of the leaf symptoms. B. Viruses under electron microscopy.
PCR detection and sequence
In order to identify the exact virus, five samples of P. multiflorum with symptoms and symptomless were tested by RT-PCR. Total RNA was extracted using the RNA-easy Isolation Reagent (Vazyme, Nanjing, China), followed by reverse transcription (RT)-PCR with pairs of universal primers for N gene of Orthotospovirusviruses (Table 1). A 777-bp amplicon was obtained from each sample (Figure 2) and cloned into the pMD18-T vector for Sanger sequencing (Takara, Dalian, China). BLASTn-analysis revealed that the 5 amplicons were identical and shared 99.20% nucleotide sequence identity with Tomato spotted wilt orthotospovirus isolate Qingdao from Tobacco (acc. no. MN861982.1). One sequence was deposited in the Gen- Bank under the accession number OP800910. Koch’s experiments also were performed and the virus from the infected plants was successfully transmitted onto healthy P. multiflorum plants (n=5) upon mechanical inoculation as the plants not only developed foliar distortion symptoms, but also tested positive for TSWV by RT-PCR with the N-specific primers (Table 1).
Figure 2:Presence of sequence in P. multiflorum plant samples by RT-PCR amplification. M: DL2000 DNA Maker; A. Tomato spotted wilt virus genus virus amplification; B. TSWV N gene fragment amplification.
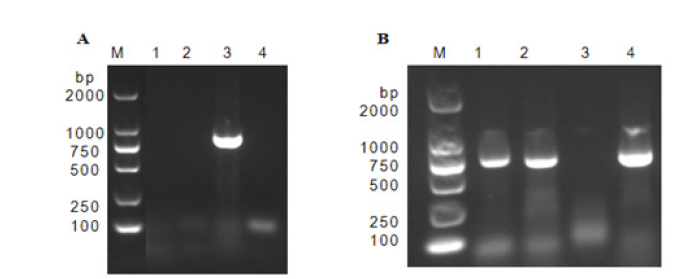
Table 1:Primers used to amplify gene sequence.
Phylogenetic analysis
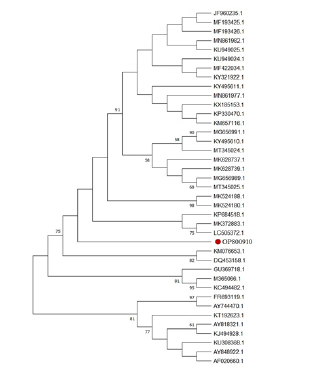
The phylogenetic tree was prepared using the available amino acid sequence of N protein of P. multiflorum and the N gene corresponding amino acid sequence of TSWV from different regions. The results are showed that the isolate of P. multiflorum grouped with several TSWV isolates (e.g., LC505372.1, MK372883.1 and KP684518.1) from Korea and South China (Figure 3).
Figure 3:A phylogenetic tree constructed based on the tomato spotted wilt virus N gene sequence
Discussion
Tomato spotted wilt orthotospovirus (TSWV), belonging to the Bunyavirus order of genus Orthotospovirus of the Tospoviridae family, is an important thrips-borne RNA virus [11]. It is the plant virus with the widest known host range, which can infect 1090 plants in 84 families, and it has caused serious economic losses to agricultural production worldwide [12]. It has been reported that TSWV can infect vegetables: Tomato and Pepper [13]; Tobacco [14], Watermelon [15], Lettuce [16]; Cardamine hirsuta and Eupatorium capillifolium [17]; Soybean [18]; Onion and Garlic [18]; Herbage: Petasites japonicus [19]; Geranium carolinianum, Gnaphalium purpureum, Linaria canadense, Molluga verticillata, Pyrrhopappus carolinianus, Raphanus raphanistrum and Triodanis perfoliate [20]; Flowers: Cardamine hirsuta and Eupatorium capillifolium [20], nasturtium [12]; Agapanthus (Bester et al., 2012); Fruits: Kiwi, Mulberry and Macadamia Nuts Fang et al. 2020 [21,22]. Even weeks can be infected [12]. In this study, the results showed that P. multiflorum with symptoms of chlorosis, mottling, leaf yellowing and necrotic in Kunming was infected with tomato spotted wilt orthotospovirus based on the data of molecular identifications and electron microscopy. This is the first report of TSWV as detected on P. multiflorum in China.
Electron microscopy (TEM) is the most visual way to detect viruses. There have been many reports on the use of TEM for virus observation. For instance, cucumber mosaic virus, tobacco rattle virus, ranunculus mild mosaic virus, zucchini tigre mosaic virus, and staphylococcus aureus were observed in different host [23-28]. In this study, the Sphular virus particles were observed under TEM by negative staining for the samples and the diameter ranged from 80 to 120nm. However, the results of TEM only identified the genus of the virus, for exact identification of the virus, we used RT-PCR detection techniques. Owing to the genome of TSWV consists of three negative-strand genomic RNA segments that are denoted large (L), medium (M), and small (S). Among them, the complementary chain of S RNA encodes the coat protein N. The N protein is an important basis for virus classification [29]. The samples of P. multiflorum infected by TSWV were tested using RT-PCR and NCBI sequence of N gene. To our knowledge, this is the first report of TSWV infected P. multiflorum in China. Symptomatic phenotype-based field surveys on some plantations in Yunnan Province indicated that the disease incidence ranged from 70% to 90%, resulting in significant loss of production of P. multiflorum. The discovery of P. multiflorum is the new host of TSWV helps to provide a new strategy for the prevention and control of the virus in the planting process [30-32].
Acknowledgement
The authors acknowledge financial support from Important plan of research and development (202103AF140003); Yunnan Fundamental Research Projects (202101AT070269).
References
- Shen X, Zhang G, Lv Q, Yang J, Wang Q, et al. (2021) Advances in chemical constituents and pharmacological activities of pleuropterus multiflorus. Journal of Tropical and Subtropical Botany 29(4): 439-450.
- Teka T, Wang L, Gao J, Mou J, Pan G, et al. (2021) Polygonum multiflorum: Recent updates on newly isolated compounds, potential hepatotoxic compounds and their mechanisms. Journal of Ethnopharmacology 271: 113864.
- Pang Z, Mao X, Xia Y, Xiao J, Wang X, et al. (2022) Multiomics reveals the effect of root rot on polygonati rhizome and identifies pathogens and biocontrol strain. Microbiology Spectrum 10(2): e0238521.
- Liao S, Chen H (1986) Polygonum multiflorum black rust and its prevention and control. TCM Bulletin pp. 13.
- Sang W, Li X, Wu W, Song B, Xiong J (2007) Indoor agent screening for the prevention and treatment of Polygonum multiflorum leaf spot disease. Pesticide pp. 60-61.
- Ciuffo M, Tavella L, Pacifico D, Masenga V, Turina M (2008) A member of a new tospovirus species isolated in Italy from wild buckwheat (Polygonum convolvulus). Arch Virol 153(11): 2059-2068.
- Margaria P, Miozzi L, Ciuffo M, Rosa C, Axtell M J, et al. (2016) Comparison of small RNA profiles in Nicotiana benthamiana and Solanum lycopersicum infected by polygonum ringspot tospovirus reveals host-specific responses to viral infection. Virus Res 211: 38-45.
- Turina M, Tavella L, Ciuffo, M (2012) Tospoviruses in the mediterranean area. Adv Virus Res 84: 403-437.
- Zhang Z, Zheng K, Dong J, Fang Q, Hong J, et al. (2016) Clustering and cellular distribution characteristics of virus particles of tomato spotted wilt virus and tomato zonate spot virus in different plant hosts. Virol J 13: 11.
- Tamura K,Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12): 2725-2729.
- Abudurexiti A, Adkins S, Alioto D, Alkhovsky S V, Avšič-Županc T, et al. (2019) Taxonomy of the order bunyavirales: Update 2019. Arch Virol 164(7): 1949-1965.
- Yu M, Yang C, Wang J, Hou Q, Zhang S, et al. (2020) First report of Tomato Spotted Wilt Virus (TSWV) isolated from nasturtium (Tropaeolum majus L) with a serious leaf mosaic disease in China. Plant Dis.
- Li Y, Tan G, Xiao L, Zhou W, Lan P, et al. (2021) A multiyear survey and identification of pepper- and tomato-infecting viruses in Yunnan province, China. Front Microbiol 12: 623875.
- Carrieri R, Sorrentino R, Lahoz E, Alioto D (2011) First report of tomato spotted wilt virus on tobacco in Campania, Italy. Plant Dis 95(5): 611.
- Pappu SS, Papp, HR, Gitaitis RD, Gay JD (1998) First report of tomato spotted wilt tospovirus infection of watermelon in Georgia. Plant Dis 82(3): 351.
- Al-Saleh MA, Al-Shahwan I M, Amer MA, Shakeel MT, Ahmad MH, et al. (2014) First report of tomato spotted wilt virus in lettuce crops in Saudi Arabia. Plant Dis 98(11): 1591.
- Groves RL, Walgenbach JF, Moyer JW, Kenned, GG (2002) The role of weed hosts and tobacco thrips, Frankliniella fusca, in the epidemiology of tomato spotted wilt virus. Plant Dis 86(6): 573-582.
- Nischwitz C, Mullis SW, Gitaitis RD, Csinos AS (2006) First Report of tomato spotted wilt virus in soybean (Glycine max) in Georgia. Plant Dis 90(4): 524.
- Kwak H R, Park G, Choi H Y, Go W R, Baek E, et al. (2020) First report of tomato spotted wilt virus in Petasites japonicus in Korea. Plant Dis.
- Stanković I, Bulajić A, Vučurović A, Ristić D, Milojević K, et al. (2012) First report of tomato spotted wilt virus infecting onion and garlic in Serbia. Plant Dis 96(6): 918.
- Meng J, Liu P, Zhu, Zou C, Li J, et al. (2015) Complete genome sequence of mulberry vein banding associated virus, a new tospovirus infecting mulberry. PLoS One 10(8): e136196.
- Wang XY, Hong Wang PG, Yang KZ, Wang PL (2016) First report of the tospovirus tomato necrotic spot associated virus infecting kiwifruit (Actinidia sp) in China. Plant Dis 100: 2539.
- Abirami R, Manoranjitham S K, Mohankumar S, Karthikeyan G (2022) Preponderance of mixed infection of cucumber mosaic virus and candidatus phytoplasma Australasia on brinjal in India. Microb Pathog 169: 105596.
- Kesharwani A K, Kulshreshtha A, Singh RP, Srivastava A, Avasthi AS, et al. (2022) First report of tobacco rattle virus infecting brassica oleracea var. Botrytis (cauliflower) in India. Plant Dis.
- Kwak HR, Byun HS, Choi HS, Kim HJ, Kim M (2022) First report of ranunculus mild mosaic virus in Ranunculus asiaticus in Korea. Plant Dis.
- Chen Y K, Shih P J, Chao HY (2022) First report of mosaic disease associated with zucchini tigre mosaic virus in wax gourd (Benincasa hispida) in Taiwan. Plant Dis.
- Yuh-Kun Chen, Pei Jung Shih, Hung Y Chao (2022) First report of mosaic disease associated with zucchini tigre mosaic virus in wax gourd (Benincasa hispida) in Taiwan. Plant Dis.
- Zeng J, Chen D, Lv C, Qin K, Zhou Q, et al. (2022) Antimicrobial and anti-biofilm activity of polygonum chinense L.aqueous extract against Staphylococcus aureus. Sci Rep 12(1): 21988.
- Zhu M, van Grinsven IL, Kormelink R, Tao X (2019) Paving the way to tospovirus infection: Multilined interplays with plant innate immunity. Annu Rev Phytopathol 57: 41-62.
- Bester R, Demas SU, Maree HJ (2023) First report of tomato spotted wilt orthotospovirus infecting agapanthus (Agapanthus praecox) in South Africa. Plant Dis.
- Fang Q, Ding M, Dong J, Yin Y, Zhang L, et al. (2013) Initial report of macadamia nuts infected with tomato spot virus. Journal of Horticulture 40(02): 350-354.
- Zhang Z, Ma X, Wu K, Zheng K, Pei W, et al. (2020) The infection cycle and epidemic characteristics of vegetable wilt in Luliang lettuce. Journal of Southwest Agriculture 33(12): 2827-2832.
© 2023 Lihua Zhao and Zhongkai Zhang. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






