- Submissions

Full Text
Modern Concepts & Developments in Agronomy
Computational Tools and Phytopathogenic Fungi Secretome: Unraveling the Protein Arsenal
Mariana Regina Almas do Carmo1, Tulio Morgan1, Rafael Ferreira Alfenas2 and Gabriela Piccolo Maitan-Alfenas1*
1Department of Biochemistry and Molecular Biology, Brazil
2Department of Phytopathology, Brazil
*Corresponding author: Gabriela Piccolo Maitan-Alfenas, Department of Biochemistry and Molecular Biology, Federal University of Viçosa, Brazil
Submission: November 02, 2022;Published: December 05, 2022

ISSN 2637-7659Volume12 Issue 1
Abstract
The industrial enzymes market is mainly segmented into carbohydrases, proteases, and lipases, being the first responsible to catalyze the depolymerization of lignocellulosic biomasses and applied to several industrial processes. Microorganisms, plant and animal tissues can be used as sources of these enzymes. However, nowadays microorganisms dominate the market as enzymes sources. The phytopathogenic fungi can secrete several carbohydrases able to degrade the host plant cell wall and important for the infection processes, which generates great appeal to produce enzymes from these fungi. Therefore, unraveling all protein arsenal secreted by a phytopathogenic fungus is interesting because these proteins may be useful for different biotechnological processes. Previous research has shown that several phytopathogenic fungi considered promising for enzymes production, such as Chrysoporthe cubensis, Ceratocystis fimbriata and Kretzschmaria zonata. In addition, computational tools have significantly contributed to these studies, allowing a better understanding of the fungal enzymatic profile and their biotechnological applications. This work shows a brief overview of the importance of understanding the enzymatic profile secreted by phytopathogenic fungi and their biotechnological applications, as well as a brief overview of computational tools applied for proteomic analysis.
Keywords:Phytopatogenic fungi; Secretome; Computational tools; Biotechnological processes
Highlights
A. Phytopathogenic fungi secrete carbohydrases of great appeal to industrial processes.
B. Secretome analysis of fungal enzymatic profiles is essential for their application.
C. Advances in fungal enzyme-based bioprocesses are possible using computational tools.
Introduction
The study of phytopathogenic fungi secretome is essential to understand the relationship between the fungus and the host plant during the infection [1]. According to [2], the secretome definition is ‘‘the global study of proteins that are secreted by a cell, tissue or organism at any given time or under certain conditions’’. By causing diseases, phytopathogenic fungi secrete enzymes for their penetration and subsequent propagation in the plant tissue. These enzymes can degrade the plant cell wall polymers, which is important for nutriente acquisition by the fungi [3,4].
Secretome analyses are necessary to obtain a global overview of the identity, function, and interaction of the arsenal of extracellular proteins that participate in plant cell wall degradation [5]. Thus, the study of phytopathogenic fungi secretome elucidate the potential role of secreted proteins involved in several metabolic processes from host infection to cell wall degradation and enables the identification and characterization of enzymes that may be applied for biotechnological processes. For instance, the comprehensive identification and quantification of the secretome of phytopathogenic strains may be a useful approach for understanding their enzymatic systems to determine new strategies for energy sources development [6].
Proteomic studies have been carried out to understand plant-fungus interactions, virulence, and fungal pathogenicity, and it is well known that there is a close relationship between virulence and the ability to secrete high levels of hydrolases in phytopathogenic fungi [6]. Furthermore, by using integrated proteomics with computational tools, it is possible to predict the subcellular location, when proteins are expressed, and the different proteins and enzymes produced by fungi. It is also possible to discover the implications for fungi development and changes in environmental responses [7-9]. Therefore, the enzymes secreted by phytopathogenic fungi deserve attention due to their potential for industrial application, which encourages research related to genetics and the secretion mechanisms of these enzymes [5].
Phytopathogenic Fungi
Fungi are heterotrophic, eukaryotic, uni or multicellular microorganisms that show a chitinous cell wall as their feature [10]. Fungi may be parasitic, saprophytic or pathogenic, and between these last classifications, some can cause diseases in plants, thus, they are denominated phytopathogenic, which can belong to several phylum such as ascomycota, basidiomycota, zygomycota and chytridiomycota [8,11]. Among the large number of identified fungal species, about 10 % can cause diseases in more than 10.000 plants, and their mode of action on the hosts can be varied, depending on the species. For example, some phytopathogenic fungi are able to invade and to colonize all plant tissues, while others act only on specific parts of the host, such as roots, stems, seeds or leaves [12].
Many fungal mechanisms and proteins have been shown to contribute to fungal pathogenicity or virulence, such as extracellular lignocellulolytic enzymes [8]. Thus, a more virulent phytopathogenic fungus shows great appeal for lignocellulolytic enzymes production and these macromolecules can be used in several biotechnological applications, such as the degradation of plant biomasses for the generation of monomeric sugars used for ethanol production [3,4,6].
Phytopatogenic Fungi Secretomes
Phytopathogenic fungi can secrete an enzymatic arsenal composed of several enzymes that act in the depolymerization of lignocellulosic biomasses, most of which are classified in the Carbohydrate-Active enZYmes (CAZy) database. The CAZy database classifies the enzymes into families and subfamilies according to their sequence, structure, and biochemical information. Enzyme classes covered by CAZy include Glycosyl Hydrolase (GH), Carbohydrate Esterase (CE), Polysaccharide Lyase (PL), Glycosyltransferases (GT), and enzymes with Auxiliary Activity (AA) [9]. It is known that CAZYmes can be applied in various industrial processes, and many studies are showing the secretion of these enzymes of commercial interest by phytopatogenic fungi [7]. The xylanases, important CAZymes secreted by several pathogen fungus can be used for various biotechnology process such as clarification of juices, pulp bleaching, bioconversion of lignocellulose into fermentable sugars and preparation of animal food and have been reported in many papers [9,13-17].
Over the years, countless research has shown the enzymatic potential secreted by phytopathogenic fungi. CAZymes and accessory enzymes produced by the pathogen fungus Penicillium funiculosum are distributed across different subfamilies (GH3, GH5, GH6, GH7, GH10, GH11, GH16, GH30, GH43, GH62, GH71, GH93, CE1, CE5, AA7 and AA9) that are responsible for biomass degradation and can be applied in several biotechnological applications [18]. Recently, our research group published data on the phytopathogenic fungi Chrysoporthe cubensis [9], Ceratocystis fimbriata [15] and Kretzschmaria zonata [16] that aroused interest in being able to secrete enzymes of great commercial appeal. Overall, studied fungi showed wide enzymatic diversity in their exoproteome, thus, the enzymes secreted by these fungi can act in several biotechnological processes, such as lignocellulosic biomass degradation and xylooligosaccharides production contributing with applicable resources in the bioenergy, food and pharmaceutical industries. Chrysoporthe cubensis (Figure 1A) is an important pathogen that can strike commercially cultivated eucalyptus species (Figure 1D) in tropical and subtropical areas of the world, which can generate losses and great damages since it causes stem canker disease of the plant species. According to [9], Chrysoporthe cubensis is capable of secreting 313 proteins, including 137 CAZymes classified as Glycosyl Hydrolases (GH), Carbohydrate Esterases (CE), Polysaccharide Lyases (PL) and Auxiliary Activities enzymes (AA). Ceratocystis fimbriata (Figure 1B) can cause damage to several agricultural and forestry crops around the world, highlighting root rot, canker, and vascular wilt (Figure 1E) in economically important species such as eucalyptus [19]. In its exoproteome, there is a β-xylosidase, an accessory enzyme of great importance in the degradation of hemicellulose that may perform an important role in the supplementation of commercial cocktails aiming to improve the yield of xylose and glucose in lignocellulosic biomass saccharification [15]. Kretzschmaria zonata (Figure 1C) is a fungus that causes root collar rot disease in teak (Figure 1F), and it was recently reported in Brazil [20]. This fungus produces an arsenal of extracellular enzymes, including xylanase, β-xylosidase and endoglucanases, enzymes with several industrial applications such as second-generation ethanol production [16].
Figure 1:Colony morphology of (A) Chrysoporthe cubensis (B) Ceratocystis fimbriata and (C) Kretzschmaria zonata on malt extract agar culture media. (D) Eucalyptus stem canker caused by C. cubensis. (E) Vascular wilt in Eucalyptus caused by C. fimbriata. (F) Root collar rot in teak caused by K. zonata.

Computational Approaches Applied to the Secretome
Proteomics, in combination with bioinformatics prediction of the secretome and enzymatic activity analyses, constitutes a powerful tool for providing information about pathogenicity, virulence factors and biotechnological potential of the exoproteome of phytopathogenic fungi [7-9]. Thus, there are many computational tools that may be employed to predict proteins of interest, their subcellular location, and conditions of expression.
To obtain bioinformatic predictions on secretome, it is necessary to obtain the organism’s genome sequence generally using high-throughput sequencing technologies, such as from Illumina and Oxford Nanopore. Thus, from sequenced genome and using appropriate tools, it is possible to unravel all arsenal of proteins secreted by an organism, opening new possibilities for their biotechnological applications. For this, the first step is the genome annotation that may be performed by several programs, such as Geneid [21], one of the first programs to predict full exonic structures of vertebrate genes in anonymous DNA sequences; GeneMark [22], a web software for gene finding in prokaryotes, eukaryotes and viruses; and Augustus [23], a gene prediction program more applied for eukaryotes, that is based on a probabilistic model of a sequence and its gene structure denominated of generalized hidden Markov model. After genome annotation, it is necessary to evaluate the quality and the confidence of the predicted gene sets and, for this, the program Benchmarking Universal Single-Copy Orthologs (BUSCO) [24] can be applied.
To predict the secreted proteins and their subcellular localization, there are several programs available, some perform simpler methods such as SECRETOOL [25] and others more refined, for example, Loctree [26], BUSCO [24] and Deeploc [27]. The programs that perform prediction refined methods can predict the subcellular location that is important in proteomics because proteins can perform a wide diversity of functions inside different cell compartments, the protein function is related to the compartment or organelle where it is located, providing a physiological context for a certain function [28]. The Loctree is a hierarchical tree system combining Support Vector Machines (SVMs) and other methods such as gene Ortology Consortium (GO) for predict the subcellular localization of proteins. It shows good levels of accuracy, but it is not used for predicting localization for membrane protein [26,29]. In contrast the Deeploc program is widely used, their prediction algorithm can differentiate several localizations including cell membrane. Their method is based on deep neural networks that do not depend on annotation of homologues from knowledge databases, relying only on sequence information. It is useful mainly for new proteins prediction that there are no annotated homologues and predict the effects of sequence variants [27].
It can also be used software capable of predicting the presence of putative signal peptides at the N-terminal of protein sequences, and secondary structures and disordered regions necessary for nonclassical secretion methods, as well as capable of postulate amino acid composition and discriminating transmembrane helices containing proteins that will be inserted into the membrane [21,30]. Therefore, the web analysis tool SECRETOOL [25] can be used for this purpose. According to [25], SECRETOOL allows the screening of different properties concerning the proteins of the fungus proteome: location of the predicted N-terminal Signal Peptide (SP), detection of the presence and location of SP cleavage sites on amino acid sequences, presence of a maximum of one Transmembrane Domain (TMD) and Glycosylphosphatidylinositol (GPI) membrane anchoring. Moreover, the proteins targeted to the nonclassical secretion pathways may be identified using other software such as Secretome P [30], a program trained on sequence features other than signaling peptides of secreted proteins, although this software is designed for bacteria and mammals and not for fungi. Thus, an alternative is an ortholog-based method to predict secreted proteins via a nonclassical pathway [30].
In phytopathogenic fungi, the CAZymes are the most important
secreted enzymes for complex carbohydrate metabolism and,
therefore, these are the proteins that generate the most commercial
interest and greater appeal for identification [9,13]. CAZymes
degrade, synthesize, and modify complex carbohydrates and
glycoconjugates in all organisms. In the protein identification step,
an approach applied to increase the CAZymes identification is using
multiple prediction strategies. For this, the predicted proteome can
be subjected to dbCAN2, a meta server for automated CAZymes
annotation based on three tools/databases:
(i) HMMER search against the dbCAN (hidden markov models)
HMMs database.
(ii) DIAMOND search against the CAZy database and
(iii) Hotpep search against the conserved CAZyme short peptide
database [31].
InterPro is a freely available database used to classify proteins sequences into families and predict domains and important sites. It is a comprehensive resource that integrates 13 protein signature databases into a single searchable resource. Sequence research performed into InterPro is powered by the InterProScan, underlying software that performs searches of proteins and nucleic acid against InterPro’s signatures [32].
Several software may be used for functional annotation such as GOanna [33], BLASTKoala [34], Blast2GO [35], and eggNOGmapper v2 [36], which are tools relying on sequence similarity for annotation. The general approach to functional annotation applied by these tools is very similar; initially, the protein sets are scanned for motifs and domains using resources like Pfam [37] and InterPro and mapped to Gene Ontology terms using GO supplied mapping files. In addition, BLAST analysis of full-length sequences may be applied to identify similar sequences which already have GO or pathway annotations linked to them and shorter motifs and domains [38].
Table 1:Software used in fungal secretome analysis.
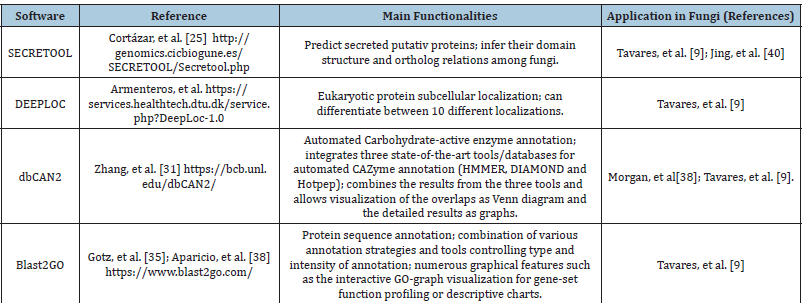
The computational tools and approaches mentioned here corroborate the phytopathogenic fungi secretome research. By properly using the available tools, it is possible to unravel the entire protein arsenal secreted by fungi, from the gene to the functional attribution of the proteins, contributing to a better understanding of the enzymatic profile of these organisms. Table 1 shows some examples of software that have been used in fungal secretome analysis [39,40].
Perspectives and Final Considerations
Over the years, research related to the phytopathogenic fungi secretome has aroused much interest because they secrete useful enzymes for biotechnological purposes such as second-generation ethanol production, food and pharmaceutical industries. The integration of computational tools, which have been improved over the years and “omics” data will allow more in-depth study of phytopatogenic fungi, enabling a greater understanding of their enzymatic profile. Therefore, new enzymes and biotechnological applications can be discovered, or applications already carried out in industrial segments can be improved. Thus, with the enzymatic profile of several fungi being unveiled, rapid advances in fungal enzyme-based bioprocesses appear to be imminent in the coming years.
References
- Fernández FJ, Carbú M, Garrido C, Vallejo I, Cantoral JM (2007) Proteomic advances in phytopathogenic fungi. Current Proteomics 4(2): 79-88.
- Hathout Y (2007) Approaches to the study of the cell secretome. Expert Reviews Proteomics 4(2): 239-248.
- Falkoski DL, Guimarães VM, Almeida MN, Alfenas AC, Colodette JL, et al. (2013) Chrysoporthe Cubensis: A new source of cellulases and hemicellulases to application in biomass saccharification processes. Bioresource Technology 130: 296-305.
- Maitan Alfenas GP, Visser EM, Alfenas RF, Nogueira BRG, Campos GC, et al. (2015) The influence of pretreatment methods on saccharification of sugarcane bagasse by an enzyme extract from Chrysoporthe Cubensis and commercial cocktails: A comparative study. Bioresource Technology 192: 670-676.
- Bouws H, Wattenberg A, Zorne H (2008) Fungal secretomes-nature’s toolbox for white biotechnology. Applied Microbiology and Biotechnology 80(3): 381-388.
- Bouwn KJ, Formolo CA, Seol H, Marathi RM, et al. (2012) Advances in the proteomic investigation of the cell secretome. Expert Reviews Proteomics 9(3): 337-345.
- Maitan AGP, Alfenas RF, Guimarães VM (2019) Phytopathogenic fungi: Useful tools to degrade plant biomass for bioethanol production. Modern Concepts & Developments in Agronomy 5(1): 499-501.
- González-Fernández R, Prats E, Jorrín-Novo JV (2010) Proteomics of plant pathogenic fungi. Journal of Biomedicine and Biotechnology 2010: 1-36.
- Tavares MP, Morgan T, Gomes RF, Rodrigues MQRB, Castro-Borges M, et al. (2021) Secretomic insight into the biomass hydrolysis potential of the phytopathogenic fungus Chrysoporthe cubensis. Journal of Proteomics 236: 104-121.
- Hawksworth DL (1991) The fungal dimension of biodiversity: Magnitude, significance, and conservation. Mycological Research 95(6): 641-655.
- Kamilov S, Mamiev M, Utaganov S, Babajanova L, Zheltenkov A, et al. (2021) Taxonomy of the mycobiota of phytopatogenic fungi of Uzbekistan. E3S web of conferences 284: 3019.
- Kubicek CP, Starr TL, Glass NL (2014) Plant cell wall-degrading enzymes and their secretion in plant-pathogenic fungi. Annu Rev Phytopathol 52: 427-451.
- Gomes KS, Maitan-Alfenas GP, Andrade LGA, Falkoski DL, Guimarães VM, et al. (2016) Purification and characterization of xylanases from the fungus Chrysoporthe cubensis for production of xylooligosaccharides and fermentable sugars. Applied Biochemistry and Biotechnology 182(2): 818-830.
- Maitan-Alfenas GP, Oliveira MB, Nagem RAP, Vries RP, Guimarães VM (2016) Characterization and biotechnological application of recombinant xylanases from Aspergillus Nidulans. International Journal of Biological Macromolecules 91: 60-67.
- Martins MP, Ventorim RZ, Coura RR, Maitan Alfenas GP, Alfenas RF, et al. (2018) The β-xylosidase from Ceratocystis Fimbriata RM35 improves the saccharification of sugarcane bagasse. Biocatalysis and Agricultural Biotechnology 13: 291-298.
- Morales ML, Almeida LF, Ladeira Ázar RIS, Guimarães VM, Alfenas RF, et al. (2021) First report on the enzymatic profile of Kretzschmaria Zonata. Biotechnology 11(9): 1-398.
- Lu L, Liu Y, Zhang Z (2020) Global characterization of GH10 family xylanase genes in Rhizoctonia Cerealis and functional analysis of xylanase RcXYN1 during fungus infection in wheat. Internation Journal of Molecular Sciences 21(5): 1812.
- Ogunyewo OA, Upadhyay P, Rajacharya GH, Okereke OE, Faas L, et al. (2021) Accessory enzymes of hipercellulolytic penicillium funiculosum facilitate complete saccharification of sugarcane bagasse. Biotechnology for Biofuls 14(1): 171.
- Harrington TC, Thorpe DJ, Alfenas AC (2011) Genetic variation and variation in aggressiveness to native and exotic hosts among Brazilian populations of Ceratocystis Fimbriata. Phytopathology 101(5): 555-566.
- Alfenas RF, Arenhart ML, Alexandre FS, Maitan-Alfenas GP (2021) Root collar rot, a new lethal disease on Tectona Grandis caused by Kretzschmaria Zonata in Brazil. Plant Dis 105(1): 221.
- Guigo R , Knudsen S, Drake N, Smith TF (1992) Prediction of gene structure. Journal of Molecular Biology 226(1): 141-157.
- Besemer J, Borodovsky M (2005) GeneMark: Web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Research 33: 451-454.
- Stanke M, Morgenstern B (2005) AUGUSTUS: A web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Research 33: 465-467.
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM (2015) BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19): 3210-3212.
- Cortázar AR, Aransay AM, Alfaro M, Oguiza JÁ, Lavín JL (2014) SECRETOOL: Integrated secretome analysis tool for fungi. Amino Acids 46(2): 471-473.
- Goldberg T, Hamp T, Rost B (2012) LocTree2 predicts localization for all domains of life. Bioinformatics 28(18): 458-465.
- Almagro JJ, Sønderby CK, Sønderby SK, Nielsen H, Winther O (2017) DeepLoc: Prediction of protein subcellular localization using deep learning, Bioinformatics 33(21): 3387-3395.
- Armengaud J, Christie-Oleza JA, Clair J, Malard V, Duport C (2012) Exoproteomics: Exploring the world around biological systems. Expert Reviews Proteomics 9(5): 561-575.
- Nair R, Rost B (2005) Mimicking cellular sorting improves prediction of subcellular localization. Journal of Molecular Biology 348(1): 85-100.
- Bendtsen JD, Jensen LJ, Blom N, Von Heijne G, Brunak S (2004) Feature-based prediction of non-classical and leaderless protein secretion. Protein Engineering Design & Selection 17(4): 349-356.
- Zhang H, Yohe T, Huang L, Entwistle S, Wu P, et al. (2018) dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Research 46(1): 95-101.
- Blum M, Chang HY, Chuguransky S, Grego T, Kandasaamy S, et al. (2021) The interpro protein families and domains database: 20 years on. Nucleic Acids Research 49(1): 344-354.
- McCarthy FM, Gresham CR, Buza TJ, Chouvarine P, Pillai LR, et al. (2011) AgBase: Supporting functional modeling in agricultural organisms. Nucleic Acids Research 39: 497-506.
- Kanehisa M, Sato Y, Morishima K (2016) BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. Journal of Molecular Biology 428(4): 726-731.
- Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, et al. (2008) High-throughput functional annotation and data mining with the Blast2GO suite, Nucleic Acids Research 36(10): 3420-3435.
- Cantalapiedra CP, Hernández-Plaza A, Letunic I, Bork P, Huerta-Cepas J (2021) EggNOG-mapper v2: Functional annotation, orthology assigments, and domains prediction at the metagenominc scale. Molecular, Biology and Evolution 38(12): 5825-5829.
- Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, et al. (2021) Pfam: The protein families database in 2021. Nucleic Acids Research 49(1): 412-419.
- Aparicio G, Götz S, Conesa A, Segrelles D, Blanquer I, et al. (2006) Blast2GO goes grid: Developing a grid-enabled prototype for functional genomics analysis. Studies in Health Technology and Informatics 120: 194-204.
- Morgan T, Falkoski DL, Tavares MP, Oliveira MB, Guimarães VM, et al. (2022) Penicillium Ochrochloron RLS11 secretome containing carbohydrate-active enzymes improves commercial enzyme mixtures during sugarcane straw saccharification. Applied Biochemistry and Biotechnology 194(7): 2946-2967.
- Jing M, Xu X, Peng J, Li C, Zhang H, et al. (2022) Comparative genomics of three Aspergillus strains reveals insights into endophytic lifestyle and endophyte-induced plant growth promotion. Journal of Fungi 8(7): 690-710.
© 2022 Gabriela Piccolo Maitan-Alfenas . This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






