- Submissions

Full Text
Modern Concepts & Developments in Agronomy
Investigation of the Cold Stress Effect on mac_4 (HSP80-Like) Gene at Transcriptional Level for Mentha × piperita L.
Selin Galatali and Ergun Kaya*
Mugla Sitki Kocman University, Faculty of Science, Molecular Biology and Genetic Department, Mugla, Turkey
*Corresponding author: Ergun Kaya, Mugla Sitki Kocman University, Faculty of Science, Molecular Biology and Genetic Department, Mugla, Turkey
Submission: March 25, 2022;Published: April 27, 2022

ISSN 2637-7659Volume10 Issue 5
Abstract
In the current study, the expression levels of mac_4 (HSP80-Like) gene, which is thought to be associated with cold stress in M. piperita, were determined by RT-PCR. First, cold stress-related genes were determined in the model organism Arabidopsis thaliana, since there is no sequence data on cold stress genes in M. piperita, and then BLAST between the two species in the NCBI database was performed to determine the sequences that matched the target genes the most. Primers specific to these sequences were also designed using the NCBI database. In order to determine the correlation of related genes with cold stress in M. piperita plant at the transcriptional level, in vitro grown plants were exposed to cold stress for 24, 72 and 168 hours. To be used in RT-PCR, cDNA was synthesized from total RNAs with reverse transcriptase enzyme. RT-PCR was performed to quantitatively detect the mRNA expression levels of target genes. RT-PCR results were analyzed by the ΔΔCT method. With 18S rRNA normalization, mRNA expression graphs of the gene were generated compared to the control group. TTEST was performed for the significance of the results, and those with p≤0.05 were considered significant. According to the TTEST result, it was observed that there was a significant increase in the mac_4 (Hsp-80 like) gene on cold treated plants.
Keywords: BLAST; Cold stress; Hsp 70; Hsp 90; mac_4 gene
Introduction
In cases of cold, disruptions are observed in biological events such as flowering and fruit formation of the plant, while the stress situation disappears when the temperature level returns to normal or the plant adapts to these temperatures [1]. In order to cope with this situation, plants try to adapt by synthesizing cryoprotectants such as biochemically soluble sugars, sugar alcohols and nitrogenous compounds or by regulating the activation of many genes that play an important role in the cold stress response such as DREB, COR, Hsp molecularly [2].
When the plant is exposed to low temperature, it first perceives this stress with changes in the membrane structure, and in this case, it causes the accumulation of calcium ions and reactive oxygen species (ROS) in the cytosol, as well as protein denaturation. Increase in calcium stimulates CaM molecule, increase in ROS stimulates TFs and activates heat shock factors (HSF). Activated HSFs enter the nucleus by translocation and bind to the heat shock element (HSE) in the target gene. With this interaction, genes such as Hsa32, ROF1, RD29A, APX2, Hsp, HSF are transcribed. As a result, the plant gains tolerance to an abiotic stress with structural and functional arrangements [3]. The current study aimed to determine the expression levels of mac_4 (HSP80-Like) gene, which is thought to be associated with cold stress in M. piperita.
Material and Methods
Plant material
Mentha x piperita L. plant was provided by the Medicinal Aromatic Plants and Local Seed Center of Mugla Metropolitan Municipality, Agricultural Department Office (Mugla, Turkey). In order to obtain the in vitro plants to be used in the study, after surface sterilization [4] of the mature material, the explants were transferred to the regeneration medium and subcultured for four-week periods at standard culture conditions [5].
Cold stres treatments
The study materials, which were subcultured in vitro and reached in sufficient numbers, were kept in the dark and at +4 °C for 24, 72 and 168 hours, covered in an opaque manner.
Molecular analysis
Since there is no study on cold stress regulation in M. x piperita, first of all, genes involved in cold stress regulation in A. thaliana were determined. Then, the target genes were determined in M. x piperita by performing BLAST over the accession number of the relevant gene or FASTA sequence in Arabidopsis thaliana plant using est and nr/nt databases in NCBI. Primers over the mRNA sequences of the heat-shock protein 80 (CV130399.1; F: 5’-CTTCTTTCACCCACTGGGCA- 3’; R: 5’-GATGCAGGTGCAAGCTGATG-3’) gene in the NCBI Primer BLAST database for later use in Q-PCR was designed.
RNA isolation was performed using the Thermo Scientific GeneJET Plant RNA Purification Kit and to be used in Real-Time PCR analysis, cDNA synthesis was performed from total RNAs with the OneScript® Plus Reverse Transcriptase OneScript® cDNA Synthesis kit. RT-PCR was also performed with AmpliqonRealQ Plus 2x Master MixGreen kit using primers designed from mRNA sequences in the NCBI Primary BLAST database in M. x piperita Determination of expression levels of genes thought to be involved in cold stress tolerance was detected in the control of reference genes. The data obtained after RT-PCR were statistically analyzed by T-Test method. A value of p≤0.05 was considered statistically significant.
Result and Discussion
Figure 1:Sequence matches as a result of BLAST between A. thaliana and M. piperita plants [HSp90, mac_4 (Hsp 80 like protein), Accesion number: CV130399/1; Match: 134/169 (%79); Gap: 5/169 (%2); Length: 290bp].

Using the NCBI database, BLAST was performed to match similar sequences in M. x piperita using sequences of desired genes in A. thaliana (Figure 1).
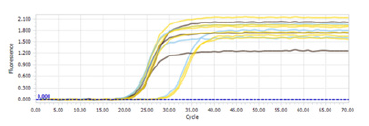
After RT-PCR, it was observed that all the peaks in the melting curve graph of the mac_4 (Hsp80-like) gene in the control and experimental groups were at different expression levels in the same time interval, and there was no primer dimer or foreign DNA contamination (Figure 2&3).
Figure 2:Melting curve plot of mac_4 (Hsp80-like) gene.
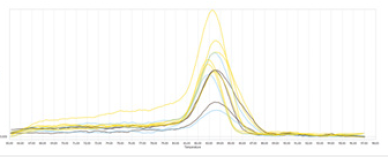
Figure 3:Amplification plot of tFighe mac_4 (Hsp80-like) gene
CT values obtained after 18S rRNA normalization with 2-ΔCT method were proportioned to the control group and a graph was created. As a result of the T-Test performed to see if the expression results are significant, there was no significant increase between the control group and the 24-hour cold stress application, while a significant increase was observed in the plants exposed to cold stress for 72 and 168 hours. While there was a significant increase between the plants exposed to cold stress for 24 hours for 72 hours and 168 hours, there was no significant difference between 72 and 168 hours (Figure 4).
Figure 4:Relative mRNA expression plot of the mac_4 (Hsp80- like) gene. *, p≤0.05; **, p≤0.01; ***, p≤0.001.
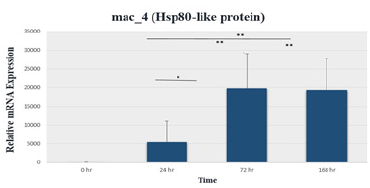
10 times upon heat shock [6]. In our study, as a result of comparing the expressions of cold stress-related genes in the control group (before cold treatment) with the 18S rRNA gene expressions in the cold acclimation period of 24, 72 and 168 hours at +4 °C, in M. x piperita, mac_4 (Hsp80-like) (Figure 4), gene expressions increased after 72 hours of incubation, and decreased after 168 hours of cold application.
Conclusion
The mac_4 gene, which is homologous to Hsp90 (A. thaliana) in M. x piperita, creates a rapid molecular response to sudden environmental factor changes that plants are exposed to [7], and increased Hsp90 transcript and protein accumulation, especially in Brassica napus, due to cold stress, suggested that they may have an important role in getting used to the cold [8]. On the other hand, the ML237 gene, which is a homolog of Hsp70 (A. thaliana) in M. x piperita, protects cells against heat and oxidative stress. It plays a very important role in the functions of preventing protein aggregation and helping proteins to refold in unsuitable environments [9-11].
References
- Lindlöf A, Chawade A, Sikora P, Olsson O (2015) Comparative transcriptomics of Sijung and Jumli Marshi rice during early chilling stress imply multiple protective mechanisms. PloS one 10(5): e0125385.
- Janska A, Marsık P, Zelenkova S, Ovesna J (2009) Cold stress and acclimation-what is important for metabolic adjustment? Plant Biology 12(3): 395-405.
- Guo M, Liu JH, Ma X, Luo DX, Gong ZH, et al. (2016) The plant heat stress transcription factors (HSFs): structure, regulation, and function in response to abiotic stresses. Frontiers in Plant Science 7: 114.
- Ozudogru EA, Kaya E, Kirdok E, Issever Ozturk S (2011) In vitro propagation from young and mature explants of thyme (Thymus vulgaris and longicaulis) resulting in genetically stable shoots. In Vitro Cellular & Developmental Biology-Plant 47(2): 309-320.
- Kaya E, Balci MA, Akguller O, Galatali S, Yeniocak S, et al. (2021) Development of an optimum proliferation medium via the graph kernel statistical analysis method for genetically stable in vitro propagation of endemic Thymus cilicicus (Turkey). Acta Botanica Croatica 80(2): 199-207.
- Nover L, Scharf KD (1984) Synthesis, modification, and structural binding of heat-shock proteins in tomato cell culture. European Journal of Biochemistry 139(2): 303-313.
- Krishna P, Gloor G (2001) The Hsp90 family of proteins in Arabidopsis thaliana. Cell Stress Chaperones 6(3): 238-246.
- Krishna P, Sacco M, Cherutti JF, Hill S (1995) Cold induced accumulation of hsp90 transcripts in Brassica napus. Plant Physiol 107(3): 915-923.
- Usman MG, Rafii MY, Martini MY, Yusuff OA, Ismail MR, et al. (2017) Molecular analysis of Hsp70 mechanisms in plants and their function in response to stress. Biotechnology and Genetic Engineering Reviews 33(1): 26-39.
- Wu CH, Caspar T, Browse J, Lindquist S, Somerville C (1988) Characterization of an HSP70 cognate gene family in Arabidopsis. Plant Physiol 88(3): 731-740.
- Duck N, McCormick S, Winter J (1989) Heat shock protein HSP70 cognate gene expression in vegetative and reproductive organ of Lycopersicon esculentum. Proceedings of the National Academy of Sciences 86(10): 3674-3678.
© 2022 Ergun Kaya. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






