- Submissions

Full Text
Modern Concepts & Developments in Agronomy
Development and Evaluation of Bioinformatics Methods a Successful Way to Design Plants for Genetic Modification
Maryam Ghasemzadeh1,3, Hamzeh Amiri1, Mahdi Khozeai2,3* and Ahmad Ismaili4
1Department of Biology, Lorestan University, Khorramabad, Iran
2Department of Biology, University of Isfahan, Isfahan, Iran
3Current address: Genetic Department, RNA Biotechnology Company, Isfahan, Iran
4Faculty of Agriculture, Lorestan University, Khorramabad, Iran
*Corresponding author: Mahdi Khozeai, Department of Biology, University of Isfahan, Isfahan, Iran
Submission: October 26, 2021 Published: February 16, 2022

ISSN 2637-7659Volume10 Issue 3
Abstract
Bioinformatics is an interdisciplinary field that combines the biology sciences, computer science and mathematics, which has gained a unique place in the life sciences. Bioinformatics has provided researchers with a very new and reliable solution for the forthcoming research in the fields of gene sequencing, genome studies and peripheral analysis, gene expression, and protomix. Another common application of bioinformatics is the identification of mononucleotide polymorphisms (SNPs) and candidate genes. Such identifications are often made with the aim of better understanding the genetic basis of diseases, matching and establishing desirable properties (especially in agricultural species), or recognizing differences between populations. Bioinformatics also seeks to better understand the structural principles of nucleic acids and protein sequences in protomix science. Gene expression studies are one of the most important characteristics that can be considered for bioinformatics, which has seen rapid growth in genetic engineering and biotechnology. For example, gene transfer with the aim of increasing expression in order to manipulate a specific biochemical pathway for the accumulation of a particular compound or metabolite, this approach can be well evaluated with the help of bioinformatics in silico environment. In this mini review, an attempt has been made to study some bioinformatics experiments, such as the phylogenetic tree and the anatomical and developmental expression of the GSA gene. This study can provide a good perspective for future laboratory studies for the researcher.
Keywords: Protomix; Bioinformatics; Environment; Phylogenetic tree
Introduction
The results of the phylogenetic tree help to identify the all-relatives gene. Actually, genetic evolution is a scientific process that allows history to be determined the evolution of groups or sequences through the trees of genetic evolution. In fact, by examining the phylogenetic tree, it is possible to discover gene function, tracing gene origin and identifying relatives one organism is provided. Phylogenetic study using molecular data such as DNA sequence for genes and amino acid sequence for proteins is very common not only in the field of evolutionary biology but also in the wide fields of molecular biology. The methods for phylogenetic study are improving along with the evolution of computer knowledge. Thus, there are many methods to infer phylogenetic tree, and many programs for each method are presented [1]. The results of the phylogenetic tree indicate less genetic distance between the members of a family.
Identifying the evolutionary relationships of a gene and recognizing the orthologous of that gene, examining the level of gene expression in different developmental and anatomical stages of the plant, determining the expression relationships between genes in addition to creating a comprehensive and clear picture of how the genome works, is a guaranty procedure for plant genetic engineering improves the desired traits. It is clear that an important and initial step in creating genetic change is study of bioinformatics and the function of the gene in differentiation and anatomical stages and how the expression of this gene is related to other genes. To do this, the first step is to use relatively new tools with much higher efficiency in comprehensive collection and analysis. There are many tools for analyzing data in public databases, one of which is Genevestigator, introduced in 2004. Genevestigator is a database and web browser with data mining capabilities for microarray data. Equipped with express analysis tools. Online analysis tools allow a wide range of information about gene expression during developmental or anatomical stages to explore patterns of gene expression. According to the website Genevestigator, the rank of a reference among all gene expression tools in the field of plant biology in all scientific journals were more than 2000 cases (related to this site).
GSA is a gene involved in ALA biosynthesis pathway. ALA is a precursor for all tetrapyrrole as example chlorophyll, heme, siroheme, Vitamin B12, phytochromobilin. These components have critical functions in living cell, as pigments (chlorophyll, phycobiliproteins), light receptor (Phytochrome), prosthetic group of many different proteins (like cytochromes, hemoglobin, myoglobin, and leghemoglobin) and enzymes (Like Catalase, Ascorbate peroxidase (or APX), Peroxidase (POD) and so on). Nowadays, ALA has received wide attention for its widespread use in agriculture, forestry and medicine. ALA at low concentrations (30-100 mg l-1) increases photosynthesis, growth, development, yield and productivity, also promoted fruit color appearance [2-4] and quality and taste of products in treated plants [4] under both normal and stressful conditions. ALA also improves nutrient uptake [5,6], antioxidant characteristics [7] and osmotic equilibrium and water use efficiency in plants. Numerous studies have shown that ALA has increased the resistance of different plants to a wide range of biotic [8] and abiotic stresses such as herbicides [9], shade [10], heat [11], cold [12], chilling [13], drought [14], salt [15], and heavy metals [16], waterlog [17], nutrient deficiency [5] and UV-B [18]. ALA also accelerates seed germination [19], and as a new plant growth regulator (PGR) and growth-promoter [20], also a potential role in tetrapyrrole-mediated retrograde signaling and exploited the direct impact of ALA biosynthesis on nuclear gene expression (NGE) [21]. ALA at high concentrations is known as an environmentally friendly and biodegradable herbicide or insecticide [22,23]. Furthermore, ALA has wild application in medical, nutrition and cosmetics [24,25], for example in cancer Photodynamic Diagnosis (PDD), Photodynamic Therapy (PDT) [26], antimicrobial drug [27] and improvement sleep [28], oral verrucous hyperplasia [29], and other clinical uses. Vitamin B12 is one of ALA byproducts. It uses as an anti-pernicious anemia factor, growth factor. ALA has sleep improvement without negative side effects like dry mouth, dizziness, continued drowsiness lasting into the next day and reminiscence problems [28]. ALA is a potential treatment for negative emotional barriers like stress and mood which various components of it are hopefulness, loneliness, and motivation [30,31]. ALA was prevention of renal injury, such as ischemia-reperfusion injury, by antioxidant effect [32]. ALA prevent chemotherapy nephrotoxicity for cisplatin without compromising the anticancer efficacy of it [33]. According to the literature data in the effect of ALA compound and its potential role in plants, which is the result of the activity of the GSA gene, it was decided to select this gene as a model for bioinformatics studies.
Database search identified one gene encoding GSA in tobacco. The microarray data available in the database (https:// genevestigator.com/gv/plant.jsp) revealed anatomical and developmental highly expression patterns for GSA gene.
Bioinformatic analysis
By blasting the GSA gene sequences at nucleotide BLAST search programs available at the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/blast/Blast. cgi), other GSA homologous sequences were found. The cladogram that shows the phylogenetic relationship between the GSA nucleotide sequences was created by using the Neighbor-Joining option present on MAGA7 software (Molecular Evolutionary Genetics Analysis) [34], Bootstrap values from 1000 were used for the creation of the cladogram. Also, determining evolutionary relationships and calculating the distance matrix by MAGA7 software.
Microarray data available through Genevestigator (https:// www.genevestigator) for GSA gene were analyzed. Genevestigator software was used for compere expression of GSA gene in two similar species of M. sativa and M. truncatula in different developmental stages and anatomical parts also survey orthologs of the GSA gene in M. truncatula and others bioinformatic analysis.
Result
Phylogenetic investigation of GSA gene
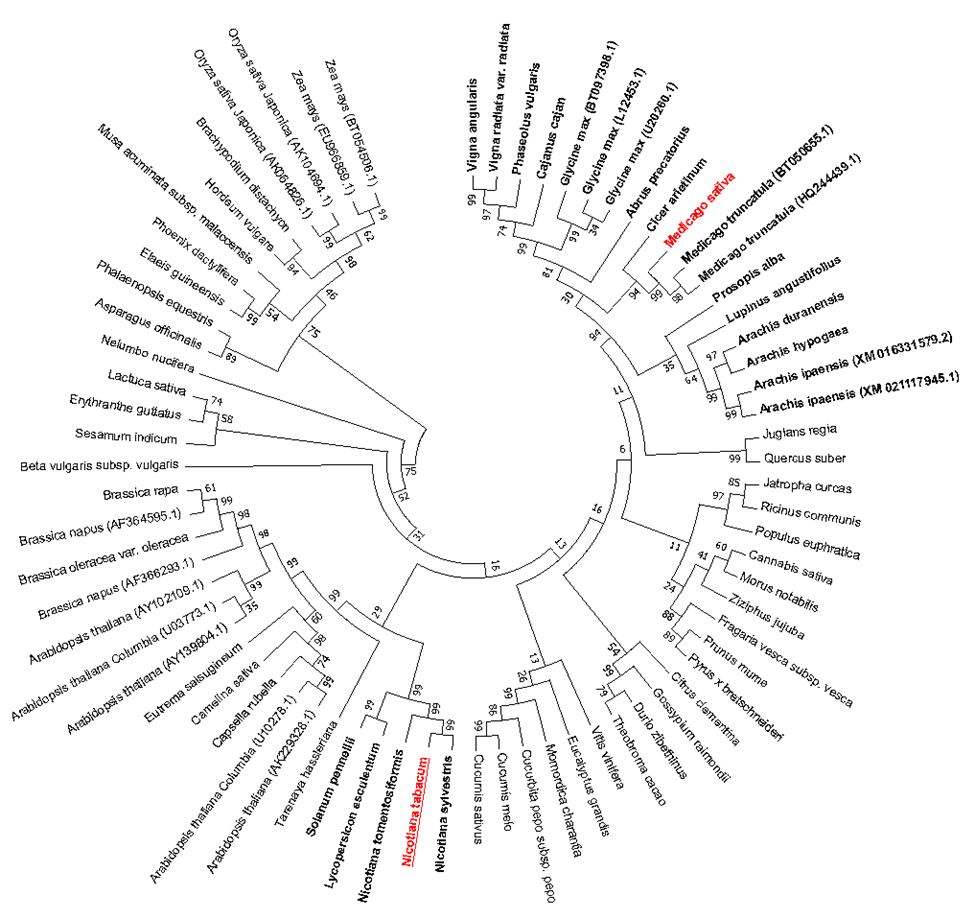
GSA gene have been found in photosynthetic organisms and in many eubacteria as no photosynthetic organisms. GSA gene and are highly conserved across all groups including higher plants and in bryophytes, cyanobacteria and many eubacteria. The phylogenetic tree was constructed based on cDNA and RNA sequences of 73 different GSA genes. As shown in Figure 1, there was indicated ten different groups of GSA gene in this phylogenetic tree which GSA gene of Medicago belongs to the Fabaceae Family. This clearly shows that Fabaceae species have more similar GSA gene than other species therefore evolutionarily are more similar to other plant groups. Also, the GSA gene of tobacco is more closely related to the GSA gene of the Solanaceae Family. Phylogenic tree was designed by MEGA7 software (Figure 1).
Figure 1: Unrooted phylogenetic tree of the GSA genes. Cladogram showing the relationship of the GSA genes in 73 terrestrial plant species. The alignments of sequences were performed using ClustalW method and grouping them using Neighbor-Joining method. Bootstrap values from 1000 pseudo-replicates were used for the creation of the cladogram and to determine the accuracy of the phylogenetic tree. phylogenic tree was designed by MEGA7. Ten different clades are illustrious among this species. Clads whose receiver and donor genes are similar indicated bolded. M. sativa as gene donor is indicated in red and Nicotiana tobacco as gene receiver is indicated in red underline.
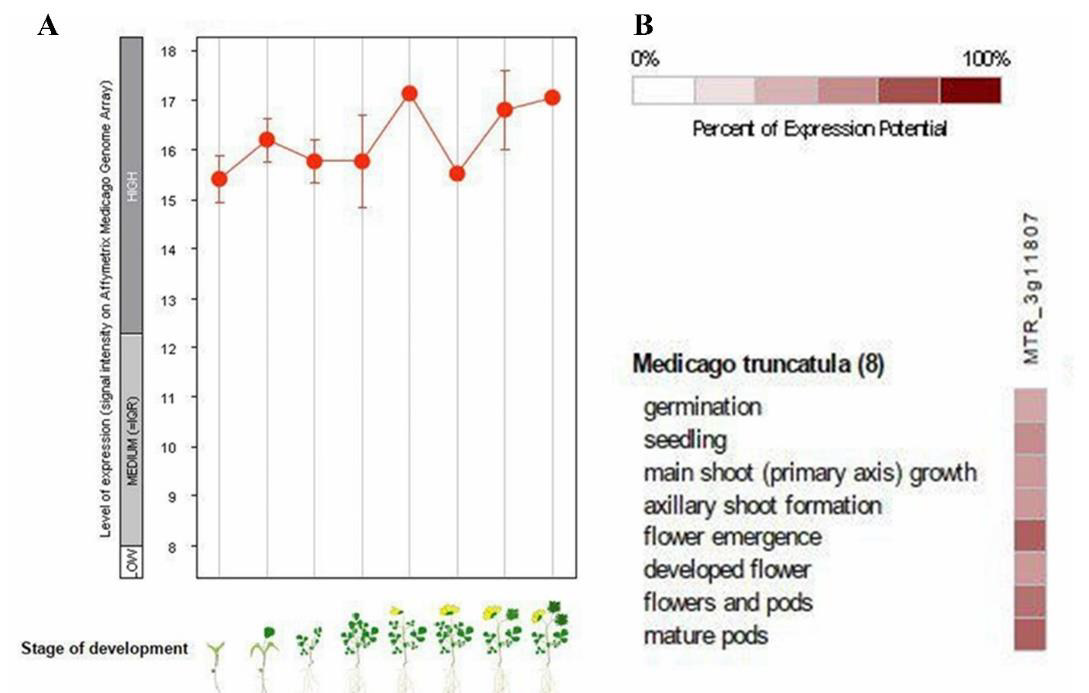
Comparison of expression of GSA gene in different developmental stage of M. truncatula
At different developmental stage in living organism each gene has different expression, and this cause differentiation [35]. Eight developmental stages of expression of GSA M. truncatula (MTR-3g118070) were studied (Figure 2). In germination stage, plant has lowest expression level among other developmental stages, about 15.4, then developed flower has a little more GSA expression. Seedling, main shoot (primary axis) growth, axillary shoot formation has medium level expression of GSA, almost 16, and highest level of expression related to flower emergence stage of plant that is about 17 then gene expression of flower and pods stage and mature pods stage are less than flower emergence stage respectively. Level of GSA gene expression in all stages of life is high, compared to expression of other genes in M. truncatula, because expression further 12 is considered high expression (Figure 2A). Percent of GSA expression potential in eight developmental stage of M. truncatula are investigated, according to Figure 2B, flower emergence, mature pods, flower and pods are exhibited 50-70 percent of gene expression potential whereas seedling, germination, main shoot growth (primary axis), axillary shoot formation, developed flower are exhibited 30-50 percent of GSA gene expression potential.
Figure 2: A. Level of GSA expression in eight developmental stages of M. truncatula. Germination, seedling,
main shoot (primary axis) growth, axillary shoot formation, flower emergence, developed flower, flower and pods,
mature pods respectively.
B. The percentage of GSA gene expression in the previous 8 steps.
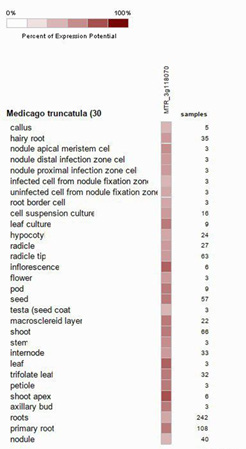
Comparison of expression of GSA gene in different anatomical parts of M. truncatula
In living organisms each gene has different expression at different anatomical parts [35]. Difference in gene expression at each group of cells cause formation of different type of cells that each one has the same genome, and this is the basis of differentiation. Thirty anatomical part of M. truncatula were investigated, results showed GSA gene expression potential in inflorescence, shoot apex, leaf and leaf culture, pod, seed, macro-sclereid layer, shoot, trifolate leaf, petiole and primary root, was about 70% to 85%, which among them inflorescence, shoot apex, leaf and leaf culture had the most GSA gene expression potential. In hairy root, nodule apical meristem cell, nodule distal infection zone cell, nodule proximal infection zone cell, uninfected cell from nodule fixation zone, root border cell, cell suspension culture, hypocotyl, radicle, radicle tip, flower, stem, internode, axillary bud expression is about 50% to 70%. In callus, infected cell from nodule fixation zone, testa (seed coat), axillary bod, roots and nodule GSA gene expression potential is about 35% to 50% which nodule, root, testa and infection cell from nodule fixation zone had lowest gene expression among them (Figure 3).
In this study, Medicago sativa was used as GSA gene donor because this species among C3 plants is very active photosynthetically) Gifford, [36] also as a main legume forage in the world due to its high nutritional value, high yield and wild adaptation [37]. So, GSA gene of Medicago sativa was candidate a good choice for study the physiological effects of increasing endogenous ALA in tobacco plant. To investigate this, first bioinformatics studies were performed, and the phylogenetic tree of this gene was studied in 73 species. This study showed the existence of 10 groups in this collection. GSA is ubiquitous in plants, existence of this gene in different families with the same function, shows that this gene plays key role in the plant, and this has led to its conservation in different species during evolution [35].
By examining the GSA gene among all plant species studied, Arabidopsis is most similar to M. truncatula in terms of sequencebased score and expression-based score. As a result, a suitable alternative for gene transfer for the overexpression of the GSA Arabidopsis gene is a more appropriate option than other species. Expressive studies of this gene at different differentiation and anatomical stages revealed that this gene is a high-expression gene in Medicago (Figure 1-3).
Figure 3: Percent of GSA expression potential in thirty anatomical stage of M. truncatula.
The most expression of GSA gene is seen in the stages of flower formation, flower pods and pod maturity because the plant is active in these stages and performs a lot of photosynthesis and therefore need to produce chlorophyll to expand photosynthetic tissues and compensate for the loss of chlorophylls that become out of the cycle. Also, the activation of mechanisms inactivation of species of ROS oxygen, for this activity of enzymes such as APX and POD and catalase is necessary, which is also present in the structure of these enzymes as a cofactor. In addition, the proteins and enzymes required by photosystem I, II pathway also have cytochromes in their structure, such as photosystem I complex and cytochrome Cytb6f, which are also essential in their function. Only in the seed germination stage, less expression of GSA gene was seen than other stages, which indicates less need for chlorophyll production and other compounds requiring heme in this stage than other developmental stages of this plant (Figures 2A&2B).
Also, in different anatomical parts, the highest relative expression of GSA gene was seen in clusters, leaves, stem tips and leaf cultures, which indicates the high requirement of these cells to synthesize compounds, proteins and enzymes that in the structure of them tetrapyrroles are necessary. while the lowest relative expression of the GSA gene is in calluses, roots, nodes, and internodes, indicating less biosynthesis of compounds, proteins, and enzymes in which tetrapyrroles are used like chlorophyll, cytochrome, catalase and APX and POD in each of these anatomical parts.
Discussion
To over expression of a gene, effort should be on the best option among other organisms. For this reason, bioinformatics data is used to help choose the best option. In this way, the expression level of the relevant gene in the source organism should be high, so that by transferring it to the destination organism, the expression level of the relevant gene is even higher than before [38,39]. To do this, one strategy is to select a gene from an organism that the sequence of the interested gene is not exactly the same as the gene of the target plant, and thus it is possible to identify and differentiate it in the plant of origin from the original gene, but at the same time this discrepancy should not be to the extent of gene expression cause conflict in the destination plant [40,41]. The second strategy, in the use of bioinformatics information is to study gene expression in computational space. In fact, the use of database information labels researcher to make the right choice in molecular programs in the laboratory by default about the desired gene or genes [42,43]. Studies of gene expression in different tissues as well as at different stages of development can reinforce the view that genetic manipulation can attain the expected achievement for the researcher. Therefore, studies showed that GSA gene of M. sativa is very high expression, therefore high expression of this gene cause increases chlorophyll concept and therefore the growth rate and biomass of M. sativa is higher than other plants [38,39]. So, in this study GSA gene of M. sativa selected as the best candidate for the source gene to transfer. As seen in the studies clearly, expression level of GSA gene in M. truncatula is high and it was included in the genes with high expression. Therefore, we used alfalfa gene to be transferred to tobacco, which has a high expression potential.
As perceived in the studies obviously, GSA gene expression is highly correlated with genes that are highly expressed during activation and development. By investigative the overexpression tobacco plant in GSA gene, higher expression of GSA was associated with higher levels of ALA content in these plants. The Molecular laboratories works also showed that tobacco GSA over expression plant had higher photosynthesis, growth rate and chlorophyll as well as elevated ALA content (Ghasemzadeh et al., not published yet).
References
- Horiike T (2016) An introduction to molecular phylogenetic analysis. Rev Agric Sci 4: 36-45.
- Xie L, Wang ZH, Cheng XH, Gao JJ, Zhang ZP, et al. (2013) 5-Aminolevulinic acid promotes anthocyanin accumulation in Fuji apples. Plant Growth Regul 69: 295-303.
- Feng X, An Y, Zheng J, Sun M, Wang L, et al (2016) Proteomics and SSH analyses of ALA-promoted fruit coloration and evidence for the involvement of a MADS-Box Gene, MdMADS1. Front Plant Sci 7: 1615.
- Ye J, Yang X, Chen Q, Xu F, Wang G, et al. (2017) Promotive effects of 5-aminolevulinic acid on fruit quality and coloration of Prunus persica (L.) Batsch. Sci Hortic 217: 266-275.
- Wei ZY, Zhang ZP, Lee MR, Sun YP, Wang LJ, et al (2012) Effect of 5-aminolevulinic acid on leaf senescence and nitrogen metabolism of pakchoi under different nitrate levels. J Plant Nutr 35(1): 49-63.
- Maruyama Nakashita A, Hirai MY, Funada S, Fueki S (2010) Exogenous application of 5-aminolevulinic acid increases the transcript levels of sulfur transport and assimilatory genes, sulfate uptake, and cysteine and glutathione contents in Arabidopsis thaliana. Soil Sci Plant Nutr 56(2): 281-288.
- Nishihara E, Kondo K, Parvez MM, Takahashi K, Watanabe K, et al. (2003) Role of 5-aminolevulinic acid (ALA) on active oxygen-scavenging system in NaCl-treated spinach (Spinacia oleracea). J Plant Physiol 160(9): 1085-1091.
- Elansary HO, El Ansary DO, Al Mana FA (2019) 5-Aminolevulinic acid and soil fertility enhance the resistance of rosemary to Alternaria dauci and Rhizoctonia solani and modulate plant biochemistry. Plants 8(12): 585.
- Xu L, Islam F, Zhang W, Ghani MA, Ali B, et al. (2018) 5-Aminolevulinic acid alleviates herbicide-induced physiological and ultrastructural changes in Brassica napus. J Integr Agric 17(3): 579-592.
- Sun YP, Zhang ZP, Wang LJ (2009) Promotion of 5-aminolevulinic acid treatment on leaf photosynthesis is related with increase of antioxidant enzyme activity in watermelon seedlings grown under shade condition. Photosynthetica 47: 347-354.
- Zhang J, Li DM, Gao Y, Yu B, Xia CX, et al. (2012) Pretreatment with 5-aminolevulinic acid mitigates heat stress of cucumber leaves. Biol Plant 56(4): 780-784.
- Wang Y, Li J, Gu W, Zhang Q, Tian L, et al (2018) Exogenous application of 5-aminolevulinic acid improves low-temperature stress tolerance of maize seedlings. Crop Pasture Sci 69(6): 587-593.
- Balestrasse KB, Tomaro ML, Batlle A, Noriega GO (2010) The role of 5-aminolevulinic acid in the response to cold stress in soybean plants. Phytochemistry 71(17-18): 2038-2045.
- Kosar F, Akram NA, Ashraf M (2015) Exogenously applied 5-aminolevulinic acid modulates some key physiological characteristics and antioxidative defense system in spring wheat (Triticum aestivum ) seedlings under water stress. South African J Bot 96: 71-77.
- Xiong JL, Wang HC, Tan XY, Zhang CL, Naeem MS, et al (2018) 5-aminolevulinic acid improves salt tolerance mediated by regulation of tetrapyrrole and proline metabolism in Brassica napus L. seedlings under NaCl stress. Plant Physiol Biochem 124: 88-99.
- Ali B, Huang CR, Qi ZY, Ali S, Daud MK, et al (2013) 5-Aminolevulinic acid ameliorates cadmium-induced morphological, biochemical, and ultrastructural changes in seedlings of oilseed rape. Environ Sci Pollut Res 20(10): 7256-7267.
- An Y, Qi L, Wang L (2016) ALA pretreatment improves waterlogging tolerance of fig plants. PLoS One 11(1): 1-15.
- Aksakal O, Algur OF, Icoglu Aksakal F, Aysin F (2017) Exogenous 5-aminolevulinic acid alleviates the detrimental effects of UV-B stress on lettuce (Lactuca sativa L) seedlings. Acta Physiol Plant 39(2): 55.
- Kanto U, Jutamanee K, Osotsapar Y, et al (2015) Promotive effect of priming with 5-Aminolevulinic acid on seed germination capacity, seedling growth and antioxidant enzyme activity in Rice subjected to accelerated ageing treatment. Plant Prod Sci 18(4): 443-454.
- Bindu RC, Vivekanandan M (1998) Hormonal activities of 5-aminolevulinic acid in callus induction and micropropagation. Plant Growth Regul 26: 15-18.
- Czarnecki O, Gläßer C, Chen JG, Mayer KFX, Grimm B, et al. (2012) Evidence for a contribution of ALA synthesis to plastid-to-nucleus signaling. Front Plant Sci 3: 236.
- Rebeiz AC, Juvik JA, Rebeiz CC (1988) Porphyric insecticides 1. Concept and phenomenology. Pestic Biochem Physiol 30(1): 11-27.
- Xu L, Zhang W, Ali B, Islam F, Zhu J, et al. (2015) Synergism of herbicide toxicity by 5-aminolevulinic acid is related to physiological and ultra-structural disorders in crickweed (Malachium aquaticum L.). Pestic Biochem Physiol 125: 53-61.
- Hunik (2002) Process for the production of vitamin b12.
- Kang Z, Zhang J, Zhou J, Qi, Q, Du G, et al (2012) Recent advances in microbial production of δ-aminolevulinic acid and vitamin B12. Biotechnol Adv 30(6): 1533-1542.
- Wachowska M, Muchowicz A, Firczuk M, Gabrysiak M, Winiarska M, et al. (2011) Aminolevulinic Acid (ALA) as a prodrug in photodynamic therapy of cancer. Molecules 16(5): 4140-4164.
- Banerjee I, Mondal D, Martin J, Kane RS (2010) Photoactivated antimicrobial activity of carbon nanotube-porphyrin conjugates. Langmuir 26(22): 17369-17374.
- Perez MH, Shintani TT, Rodriguez BL, Davis J, Harrigan RC, et al. (2013) The role of 5-aminolevulinic acid (5-ALA) and sleep. Int J Clin Med 04(10A): 1-7.
- Chen H, Chen C, Yang H, Kuo MYP, Kuo YS, et al. (2004) Successful treatment of oral verrucous hyperplasia with topical 5-aminolevulinic acid-mediated photodynamic therapy. Oral Oncol 40(6): 630-637.
- Suzuki H, Masuki S, Morikawa A, Ogawa Y, Kamijo YI, et al. (2018) Effects of 5-aminolevulinic acid supplementation on home-based walking training achievement in middle-aged depressive women: randomized, double-blind, crossover pilot study. Sci Rep 8(1): 7151.
- Aquino R, Perez M, Sil P, Shintani T, Harrigan R, et al. (2018) The relationship of 5-Aminolevulinic acid on mood and coping ability in prediabetic middle aged and older adults. Geriatrics 3(2): 17.
- Uchida A, Kidokoro K, Sogawa Y, Itano S, Nagasu H, et al (2019) 5-Aminolevulinic acid exerts renoprotective effect via Nrf2 activation in murine rhabdomyolysis-induced acute kidney injury. Nephrology 24(1): 28-38.
- Terada Y, Inoue K, Matsumoto T, Ishihara M, Hamada K, et al. (2013) 5-Aminolevulinic acid protects against cisplatin-induced nephrotoxicity without compromising the anticancer efficiency of cisplatin in rats in vitro and in vivo. PLoS One 8(12): 1-12.
- Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7): 1870-1874.
- Ojolo SP, Cao S, Priyadarshani SVGN, Li W, Yan M, et al. (2018) Regulation of plant growth and development: a review from a chromatin remodeling perspective. Front Plant Sci 9: 1232.
- Gifford RM (1974) A comparison of potential photosynthesis, productivity and yield of plant species with differing photosynthetic metabolism. Aust J Plant Physiol 1: 107-117.
- Cao Y, Zhang Z, Zhang T, You Z, Geng J, et al (2018) Overexpression of a zeaxanthin epoxidase gene from Medicago sativa enhances the tolerance to low light in transgenic tobacco. Acta Biochim Pol 65(3): 431-435.
- Spiga E, Degiacomi MT, Dal Peraro M (2014) New strategies for integrative dynamic modeling of macromolecular assembly, (1st edn), Elsevier Inc.
- Wong KC (2021) Computational biology and bioinformatics. CRC Press.
- Dawson WK, Maciejczyk M, Jankowska EJ, Bujnicki JM (2016) Coarse-grained modeling of RNA 3D structure. Methods 103: 138-156.
- Kmiecik S, Gront D, Kolinski M, Wieteska L, Dawid AE, et al. (2016) Coarse-grained protein models and their applications. Chem Rev 116(14): 7898-7936.
- Sim A, Minary P, Levitt M (2014) Modeling nucleic acids. Curr Opin Struct Biol 22(3): 273-278.
- Joyce AP, Zhang C, Bradley P, Havranek JJ (2015) Structure-based modeling of protein: DNA specificity. Brief Funct Genomics 14(1): 39-49.
© 2022 Mahdi Khozeai. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






