- Submissions

Full Text
Modern Concepts & Developments in Agronomy
Genetics of Seed-Protein Content and Yield Related Traits in Pea (Pisum sativum L.)
M M Uzzal Ahmed Liton*, Md Harunur Rashid and Md. Ehsanul Haq
Department of Plant Science, 222 Agriculture Building, University of Manitoba, Winnipeg, Manitoba R3T2N2, Canada
*Corresponding author:M M Uzzal Ahmed Liton, Department of Plant Science, 222 Agriculture Building, University of Manitoba, Winnipeg, Manitoba R3T2N2, Canada
Submission: June 01, 2021Published: June 28, 2021

ISSN 2637-7659Volume9 Issue 1
Abstract
Grain legumes are important for food security and nutritional value. They produce protein and protein rich foods for millions of people around the globe. They currently play a central role in sustainable agricultural production system by adding nitrogen nutrient to the soil for the subsequent crops largely through symbiosis with nitrogen-fixing soil bacteria. This sustainable provision of nitrogen fertilizer allows reducing the fertilization cost in cropping system and greenhouse gas emission. Pea (Pisum sativum L.) is one of the most important grain legumes which produce higher amount of seed protein and provides solution for protein deficiency and protein choices. It is also a good source of plant-based protein and has high nutritional value for human and animal consumption. With the increasing demand of plant-based protein, increasing pea seed yield and seed protein content (SPC) are the most important breeding targets today. Therefore, it is needed to elucidate the existing literature regarding genetics of pea. Here we review the existing potential quantitative and qualitative genetics underlying SPC and yield attributes in pea, currently used methods in pea breeding, and future direction for sustainable production.
Introduction
Protein-rich plant-based foods reduce detrimental health effects of millions of mothers and their children in low animal protein intake regions in the world [1]. In contrast, animalbased protein food sufficient countries such as Canada, USA, and UK are moving towards plantbased protein which creates a lot of pressure on crops with high protein content such as peas. Field pea (Pisum sativum L.) is one of the most important protein-rich legumes with some other agronomic benefits. It is an important green vegetable and the second most important grain legumes worldwide after common bean [2]. It provides dietary protein for millions of people globally [3]. Field pea also has some agronomic benefits such as nitrogen fixation through symbiosis with nitrogen-fixing bacteria resulting less use of nitrogen fertilizer in cropping system [4]. Due to these benefits field pea considers as a good source of protein and resources for farmers in marginal environments. Improving seed protein content (SPC) along with increased seed yield in dry peas is important to alleviate malnutrition in the world and facilitate moving towards plant-based protein food industry. However, a limited amount of research has been carried out on the genetics of SPC and its relation to beneficial agronomic traits for seed yield.
Pea is a cool-season grain legume belongs to Leguminosae family (Genus: Pisum, subfamily: Faboideae tribe: Fabeae). It is cultivated worldwide for both grains and vegetables with differences in morphology and nutritional status [5]. It is a self-pollinated diploid species (2n=2x =14) and its genome size (4300Mb) is 10 times larger than the model species Medicago truncatula (500Mb). This expanded genome size in peas constitutes higher amount of repetitive DNA composing various families of mobile genetic elements [6]. In contrast, the exotic portion of genome is much lower in proportion to the whole genome compared to other legume species such as M. truncatula, Lotus aponicas (c. 472Mb) and chickpea (Cicer arietinum L.) (c. 740Mb). However, correlated and genetically programmed processes associated with SPC, and other agronomic traits have been reported in previous studies on legumes. A little is known about the molecular genetics in pea which limits the identification of favourable alleles and trait improvement through marker-assisted selection (MAS) breeding. The invention of high-throughput research tools and reducing cost of next generation sequencing provide durable solution to dissect the genetic architecture of model and non-model legume species [3,7]. Therefore, we can be able to bridge the knowledge gap between models and crops for trait improvement which is greatly applicable for improving SPC and yield-related agronomic traits in pea.
Quantitative genetics
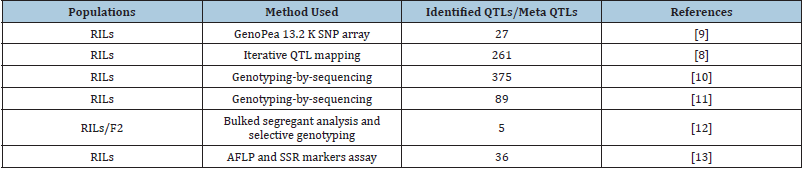
Genetic parameters associated with quantitative traits such as number of loci, effect and sizes can be estimated through QTL analysis. Most of the parameters can be population specific resulting differences in identification of QTLs in different population. However, QTL analysis preceded by multiple populations in multienvironments is very common in genetic studies to determine stable QTL and correlation among traits. SPC and seed yield are complex traits and inherited mostly quantitatively. Burstin et al. [8] reported that QTLs associated with SPC, and seed yield are either consistent with plant source capacity controlling genes for production and fulfillment of seeds or seed sink strength controlling genes involved in synthesis of storage product and seed formation. Several loci associated with yield attributes and SPC has been identified in pea [8-13] (Table 1) and is currently being utilized in the respective research. However, the genetics basis of SPC and agronomic traits in peas is needed to be uncovered by more collaborative research worldwide.
Table 1: Detected QTLs associated with seed-protein content (SPC) and yield related traits in recombinant inbreed lines (RILs) of pea.
Qualitative genetics
A few genes and markers associated with yield attributes and SPC has been identified in pea [8,14-17] (Table 2) and is currently being utilized in the variety development programmes. Starch- Branching Enzyme 1 of r locus and ADP glucose-phosphorylase of rb locus are known to control wrinkled seed phenotype and impact seed development, yield and protein content [14,15]. Single gene such as Le, Af and rugosus have reported previously which are involved in controlling plant architecture such as internode length, leaf type and seed size [8]. A subtilase gene SBT1.1 controlling seed size by providing molecules that can act as signals to control cell division within the embryo [16]. Gali et al. [17] reported SNP markers linked to plant height, grain yield, seed starch and seed protein concentration in different chromosomes in a genome wide association study (GWAS), which have potential for marker-assisted selection towards rapid cultivar improvement.
Table 2: Identification of major genes/markers link to seed-protein content and yield related traits in pea.

Concluding remarks and future perspective
Identification of causative polymorphism for SPC and yield attributes has been challenging in pea. Low resolution of classical quantitative genetics to dissect genetic architecture of SPC and agronomic traits in model and non-model species limits the use of this approach. Similarly, information about the genetic basis of relationship between SPC and agronomic traits like seed yield, seed weight, days to flowering and growth habit are limited in pea. Therefore, it is essential to determine the genetic basis of trait correlation in pea for improving and stabilizing SPC while maintaining yield and other agronomic attributes. The future research directions are as follows:
A. Develop mapping population and breeding lines utilizing available and promising germplasms.
B. Utilize recently advanced technologies such as SNP, NGS etc. in breeding program to develop varieties with improved quality.
C. Develop and execute research programmes towards identification of new QTLs/genes using all available germplasm globally.
D. Develop new pea varieties and improve existing varieties based on different agro-ecological zones.
E. Perform a meta-analysis of research on global pea production systems.
F. Develop the best agronomic practices for pea based on different agro-ecological zones.
G. Develop diverse pea cropping systems for increased the durability of existing QTLs/locus/genes.
H. Develop integrated management systems that allow for increased the effectiveness of genetics and lowered the use of biocides.
I. Evaluate the efficacy of tools and methods particularly large data analysis to improve genetic pool in the existing pea germplasm.
References
- Li L, Zheng W, Zhu Y, Ye H, Tang B, et al. (2015) QQS orphan gene regulates carbon and nitrogen partitioning across species via NF-YC interactions. Proc Natl Acad Sci 112(47): 14734-14739.
- Kreplak J, Madoui MA, Cápal P, Novák P, Labadie K, et al. (2019) A reference genome for pea provides insight into legume genome evolution. Nat Genet 51(9): 1411-1422.
- Varshney RK, Chen W, Li Y, Bharti AK, Saxena RK, et al. (2012) Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat Biotechnol 30: 83-89.
- Akibode CS, Maredia M (2012) Global and regional trends in production, trade and consumption of food legume crops. Department of Agricultural, Food, and Resource Economics, Michigan State University, USA.
- Liu N, Zhang G, Xu S, Mao W, Hu Q, et al. (2015) Comparative transcriptomic analyses of vegetable and grain pea (Pisum sativum) Seed Development. Front Plant Sci 6: 1039.
- Macas J, Neumann P, Navrátilová A (2007) Repetitive DNA in the pea (Pisum sativum) genome: comprehensive characterization using 454 sequencing and comparison to soybean and Medicago truncatula. BMC Genomics 8: 427.
- Obala J, Saxena RK, Singh VK, Kale SM, Garg V, et al. (2020) Seed protein content and its relationships with agronomic traits in pigeonpea is controlled by both main and epistatic effects QTLs. Sci Rep 10(1): 214.
- Burstin J, Marget P, Huart M, Moessner A, Mangin B, et al. (2007) Developmental genes have pleiotropic effects on plant morphology and source capacity, eventually impacting on seed protein content and productivity in pea. Plant Physiol 144(2): 768-781.
- Klein A, Houtin H, Rond Coissieux C, Naudet Huart M, Touratier M, et al. (2020) Meta-analysis of QTL reveals the genetic control of yield-related traits and seed protein content in pea. Sci Rep 10: 15925.
- Gali KK, Liu Y, Sindhu A, Diapari M, Shunmugam ASK, et al. (2018) Construction of high-density linkage maps for mapping quantitative trait loci for multiple traits in field pea (Pisum sativum). BMC Plant Biol 18(1): 172.
- Ma Y, Coyne CJ, Grusak MA, Mazourek M, Cheng P, et al. (2017) Genome-wide SNP identification, linkage map construction and QTL mapping for seed mineral concentrations and contents in pea (Pisum sativum). BMC Plant Biol 17(1): 43.
- Timmerman Vaughan GM, McCallum JA, Frew TJ, Weeden NF, Russell AC (1996) Linkage mapping of quantitative trait loci controlling seed weight in pea (Pisum sativum). Theor Appl Genet 93: 431-439.
- Ubayasena L, Bett K, Taran B, Warkentin T (2011) Genetic control and identification of QTLs associated with visual quality traits of field pea (Pisum sativum). Genome 54(4): 261-272.
- Bhattacharyya MK, Smith AM, Ellis THN, Hedley C, Martin C (1990) The wrinkled-seed character of pea described by Mendel is caused by a transposon-like insertion in a gene encoding starch-branching enzyme. Cell 60(1): 115-122.
- Smith AM, Bettey M, Bedford ID (1989) Evidence that the rb locus alters the starch content of developing pea embryos through an effect on ADP Glucose Pyrophosphorylase. Plant Physiol 89(4): 1279-1284.
- D’Erfurth I, Le Signor C, Aubert G, Sanchez M, Vernoud V, et al. (2012) A role for an endosperm-localized subtilase in the control of seed size in legumes. New Phytol 196(3): 738-751.
- Gali KK, Sackville A, Tafesse EG, Lachagari VBR, McPhee K, et al. (2019) Genome-wide association mapping for agronomic and seed quality traits of field pea (Pisum sativum). Front Plant Sci 10: 1538.
© 2021 M M Uzzal Ahmed Liton. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and build upon your work non-commercially.
 a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in
a Creative Commons Attribution 4.0 International License. Based on a work at www.crimsonpublishers.com.
Best viewed in 







.jpg)






























 Editorial Board Registrations
Editorial Board Registrations Submit your Article
Submit your Article Refer a Friend
Refer a Friend Advertise With Us
Advertise With Us
.jpg)






.jpg)














.bmp)
.jpg)
.png)
.jpg)










.jpg)






.png)

.png)



.png)






